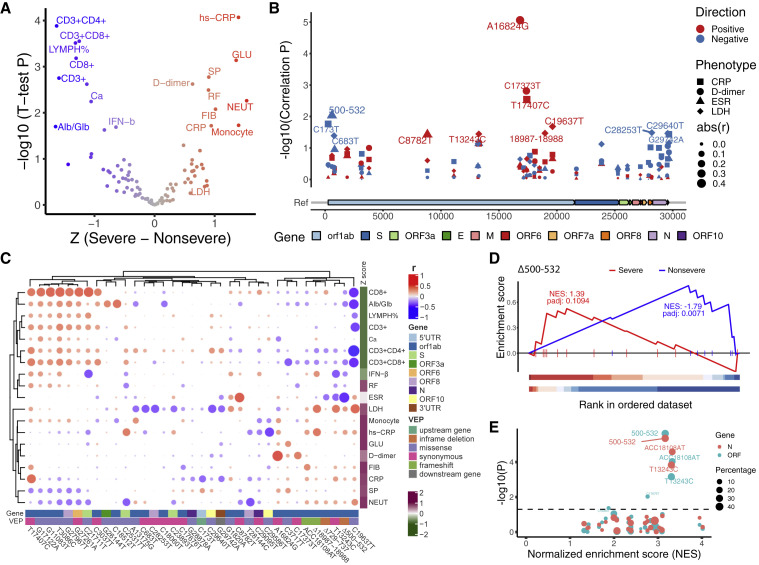
Figure 3.
Associations of 35 recurrent genetic variants with clinical phenotypes
(A) Volcano plot of the significant traits that are associated with COVID-19 severity. The x axis shows the difference of Z score between patients that developed severe or non-severe symptoms. The y axis shows –log10(p value) of t test between these two groups. Red indicates indicating severe related traits, and blue, non-severe related traits. Severe related traits include CRP (C-reactive protein), hs-CRP (high-sensitivity CRP), GLU (serum glucose), SP (systolic pressure), D-dimer, RF (respiratory frequency), FIB (fibrinogen), NEUT (neutrophils counts), Monocyte (monocyte counts), and LDH (lactate dehydrogenase); and non-severe related include CD3+, CD3+CD4+, CD3+CD8+, CD8+ cell counts, LYMPH% (percentage of lymphocytes), Ca (serum calcium), Alb/Glb (albumin and globulin ratio), and IFN-b (serum IFN-β). The traits with p value less than 0.05 are shown.
(B) Manhattan plot of 35 genetic variants that were associated with CRP, D-dimer, ESR (erythrocyte sedimentation rate), and LDH. Genome position and –log10 (correlation p value) were shown. Different symbols indicate that correlated phenotypes and size of the symbols is in proportion to the absolute (abs) value of correlation efficient (r). Red or blue indicates a positive or negative correlation, respectively.
(C) Heatmap of correlation coefficients between clinical phenotypes and genetic variants. The clustering was based on Pearson correlation coefficients. Intensity of red and blue indicates correlation efficient, red indicates positive correlation, and blue, negative correlation. Intensity of purple and green indicates higher or lower mean values in the severe group.
(D) Δ500-532 is significantly enriched with non-severe traits but not with severe traits. p values were adjusted using permutation.
(E) Dot plot shows summed normalized enrichment score (NES) and p values of t test between the group with or without a certain variant for Ct values in qPCR tests. The dots in the top right corner have more significant p values for Ct values of N (red) or ORF (blue) genes and a higher enrichment score.