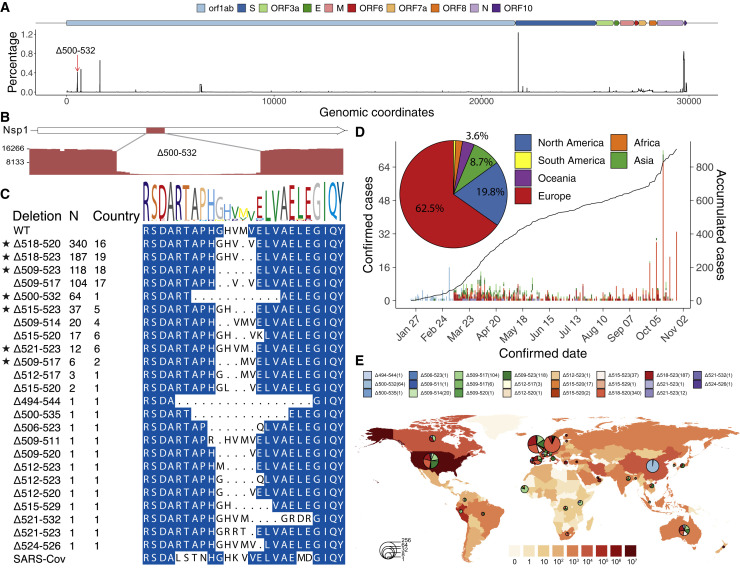
Figure 4.
Prevalence of deletion variants in the 500-532 locus in SARS-CoV-2 genome
(A) Percentage of deletion in the SARS-CoV-2 genome. The red arrow indicates the 500-532 locus.
(B) Sashimi plot indicates the sequencing reads that were mapped around the identified Δ500-532 region (see Figures S6B and S6C).
(C) The alignment of amino acid residues of mutation neighboring 500-532 locus in Nsp1 coding region. Identified cases number (N) and the number of countries were shown. WT, wild-type SARS-CoV-2 Nsp1 sequence. Stars indicate the deletion mutants that were further tested experimentally (related to Figures 5, 6, and 7).
(D) The confirmed (left y axis) or accumulated (right y axis) cases with Nsp1 deletion mutants. The pie chart shows the percentages of cases identified in each continent.
(E) Geographic distribution of COVID-19 cases with the deletion variants worldwide. Color intensity on the map indicates confirmed cases in that region. The pie charts indicate the proportions of different deletion variants in each country, and the size of pie charts indicate the number of cases with the deletion mutants.