Figure 1.
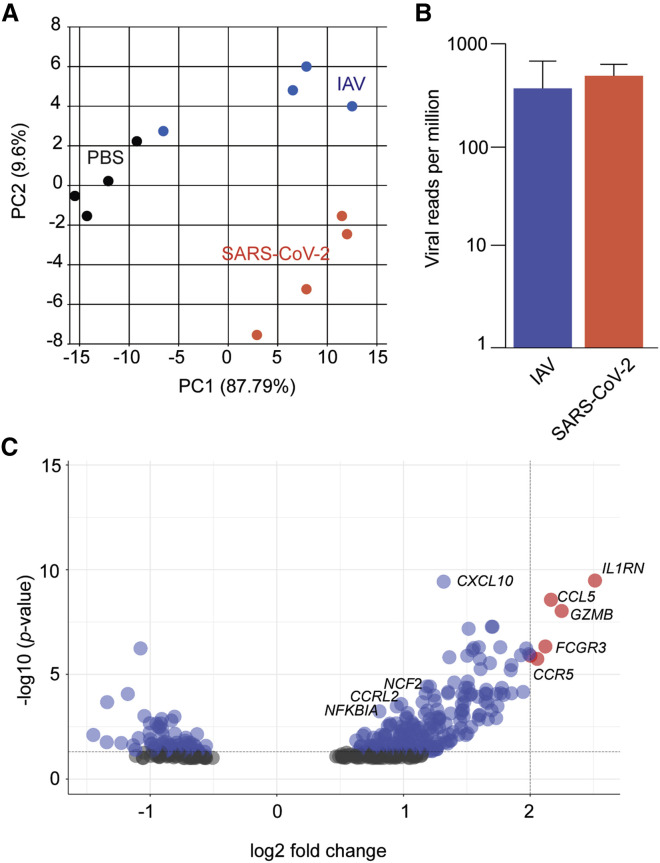
The transcriptional signatures from SARS-CoV-2- and IAV-infected hamster lungs differ
(A) Principal-component analysis of mRNA-seq samples derived from total lungs of hamsters (n = 4/cohort) treated with IN PBS, IAV (A/Cal/04/2009), or SARS-CoV-2 (USA-WA1/2020), collected 5 days after treatment, and aligned to virus and hamster reference genomes.
(B) Reads per million of SARS-CoV-2 and IAV from (A) relative to host mRNA. Error bars indicate standard error of the mean.
(C) Volcano plot comparing DEGs following SARS-CoV-2 versus IAV infection as described in (A) (gray, not significant; blue, significant; red, significant and log2 fold change > 2). Genes are called by their human ortholog.
See also Table S1.