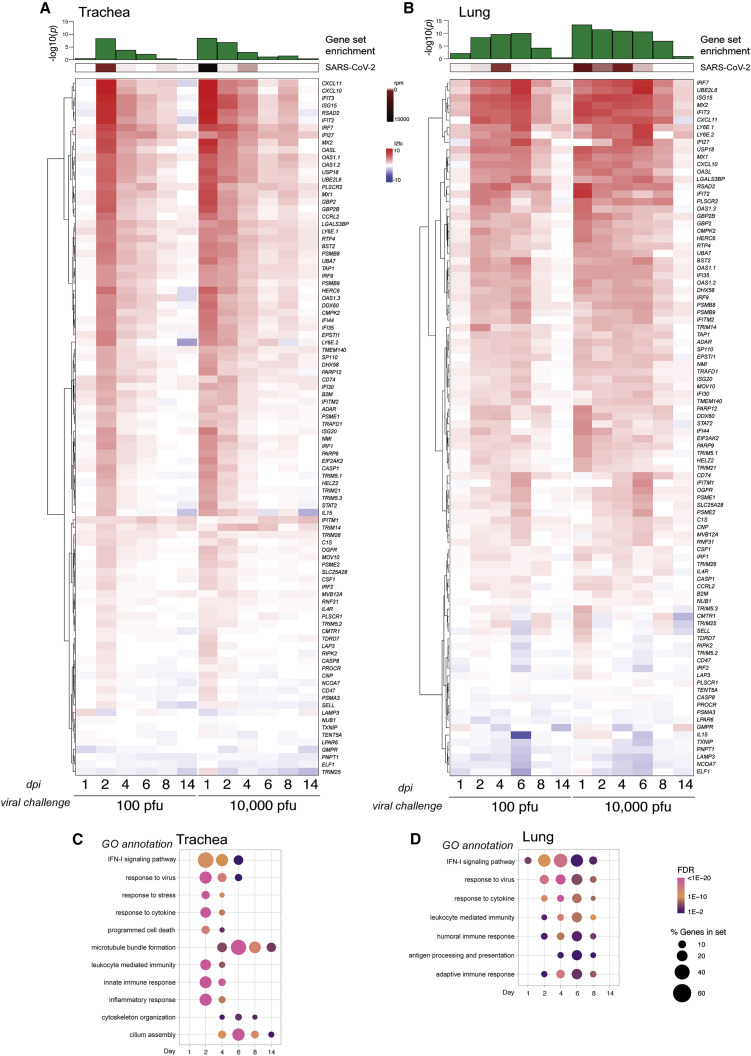
Figure 4.
The upper and lower respiratory tract IFN-I transcriptional signatures
(A and B) Differential gene expression of a curated list of ISGs calculated from bulk mRNA-seq from (A) tracheae and (B) lungs of hamsters infected with SARS-CoV-2 for the times and inocula indicated compared with uninfected controls. The heatmaps represent the log2 fold change of each gene indicated on the right (human ortholog) at the time points below. Averages of reads per million (rpm) mapping to the SARS-CoV-2 genome in both tissues throughout the time course are shown above each heatmap (in brown), and the statistical significance of the enrichment of the gene list in the total number of DEGs (in green) is shown as −log10(adjusted p value) (control group, n = 5; each experimental group, n = 3).
(C and D) Gene enrichment analysis of all DEGs (log2 fold change > 1, false discovery rate [FDR] < 0.05) increased during infection for (C) the trachea and (D) the lungs, as depicted for the time course of infection with 100 PFU. Each dot indicates a statistically significant representation of the category of genes listed on the left at each time point. The size of the dot indicates the percentage of enriched genes from each GO annotation set, and the color represents its FDR.
See also Figure S4.