Figure 6.
Sites distal to direct SARS-CoV-2 replication display significant inflammation
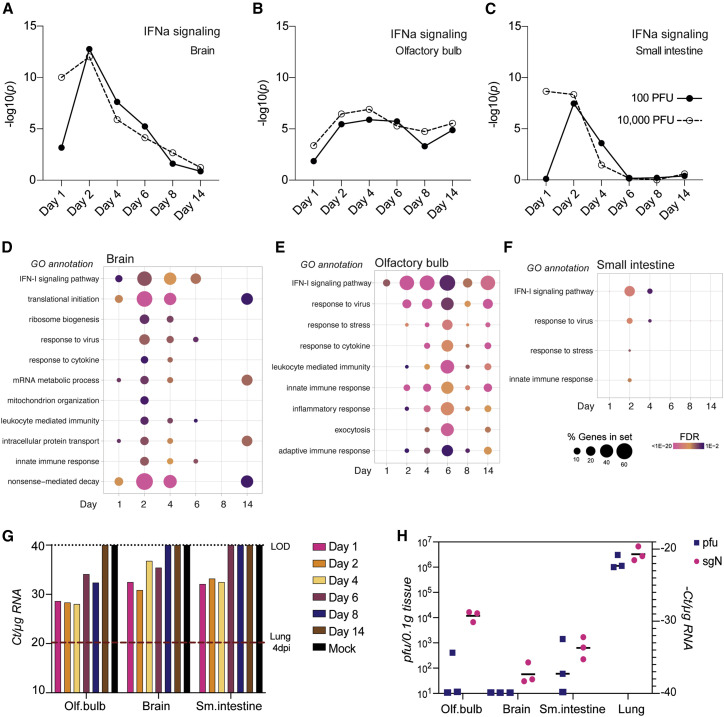
(A–C) The statistical significance of the enrichment of a curated list of ISGs among the total number of DEGs is shown as –log10(adjusted p value) above each time point from transcriptome analysis of total RNA extracted from (A) the brain, (B) the olfactory bulb, or (C) the small intestine (all groups n = 3; small intestine 10,000 PFUs day 8, n = 2).
(D and E) Gene enrichment analysis of all DEGs (log2 fold change > 1, FDR < 0.05) that increased during infection in (D) the brain, (E) the olfactory bulb, and (F) the small intestine are depicted for the time course of infection with 100 PFU. Each dot indicates a statistically significant representation of the category of genes listed on the left at each time point. The size of the dot indicates the percentage of enriched genes from each GO annotation set, and the color represents its FDR (n = 3).
(G) sgN levels represented by real-time qRT-PCR Ct score per 1 μg RNA from the olfactory bulb, brain, and small intestine at the indicated time points after infection with 100 PFUs SARS-CoV-2. RNA samples of each organ and corresponding time point were pooled before analysis (n = 3 per condition). The red dotted line indicates the Ct value of a sample corresponding to lungs collected from a hamster 4 days after infection.
(H) sgN levels represented by real-time qRT-PCR (−)Ct score per 1 μg RNA and PFUs per 100 mg of analyzed tissue homogenate (n = 3 per condition). Horizontal bars indicate mean.