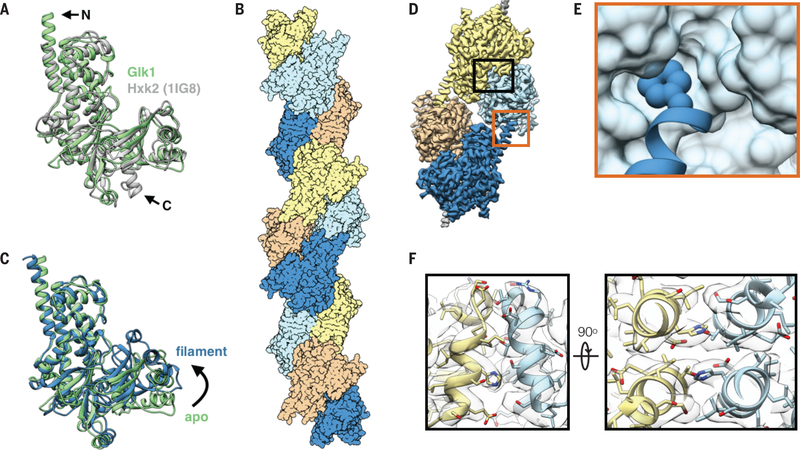
Fig 2. Glk1 forms anti-parallel, two-stranded filaments in its closed state.
A: Superimposition of Glk1 crystal structure (green) (residues 1–500) with Hxk2 (PDB ID: 1IG8) (19) (white) (residues 18–486). The N-terminal helix of Glk1 extends further than that of Hxk2 (arrow, N), while the C-terminal helix of Hxk2 extends further (arrow, C). B: Surface representation of a model of Glk1 filaments reconstructed from cryo-EM (3.8 Å resolution). Glk1 filaments are two-stranded, anti-parallel helices. Subunits along each strand are either orange/yellow or blue/cyan. C: Superimposition of the Glk1 crystal structure (green) with the Glk1 filament conformation from the cryo-EM reconstruction (blue). The crystal structure is not ligand bound and is in the open state while the filament form is ligand bound and in the closed state. D: Cryo-EM map of four subunits in the Glk1 filament colored by subunit. Longitudinal contact is boxed in orange, and a lateral contact is boxed in black. E: Closeup of longitudinal contact with Phe3 represented as van der Waals spheres and the next subunit represented as a surface model. Phe3 of one subunit inserts into the hydrophobic pocket near the next subunits C-terminus. F: Two orthogonal close-ups of lateral filament contact. The helix-loop-helix from residue 371–393 of one subunit (yellow) binds antiparallel to the same region of the adjacent subunit (blue). Cryo-EM map is transparent grey.