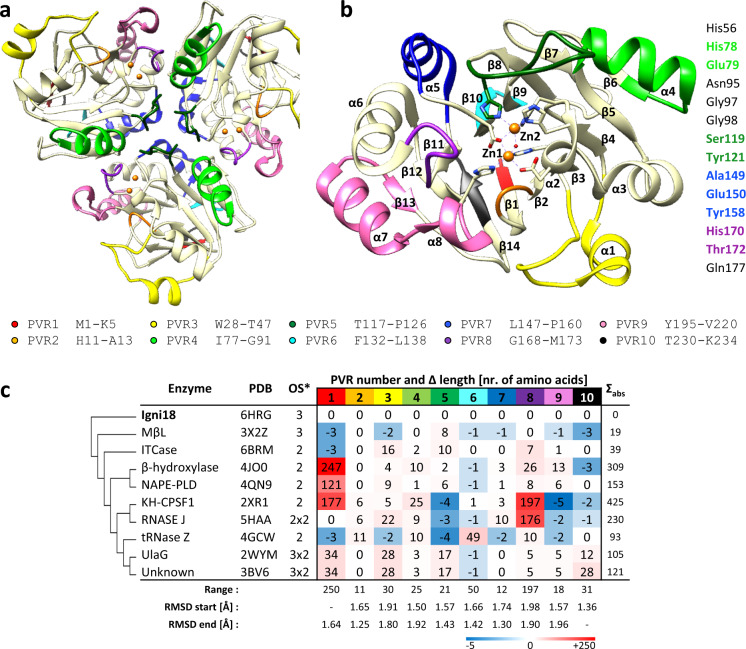
Fig. 1. The crystal structure of Igni18 reveals the origin of diverse MβLs.
The protein crystalized as a homotrimer (a). A monomer (b) comprises 8 α-helixes and 14 β-strands and contains two Zn2+ ions (orange). Ten Protein Variable Regions (PVRs) describe structural evolution and specialization within the MβL family and are depicted in a and b. The amino acids comprising each PVR are indicated underneath. In total, 9 out of 14 amino acids needed for stabilization of the trimer are found within PVRs 4, 5, 7, and 8 (light-green, dark-green, blue, and purple; right), leading to an evolutionary fast loss of this quaternary structure. Differences in the number of amino acids of the described PVRs among 10 MβL-fold-containing structures (c); Molecular relation (left) and absolute variation in the number of amino acids (Σabs, right) compared to Igni18 as well as statistic data on PVR descriptors (total length range > 10 amino acids, start/end position RMSDs for all aligned structures <2 Å; bottom) are also given. Asterisk represents Oligomerization state of the crystallized enzyme (3: trimer; 2: dimer; 2 × 2: double dimer; 3 × 2: triple dimer).