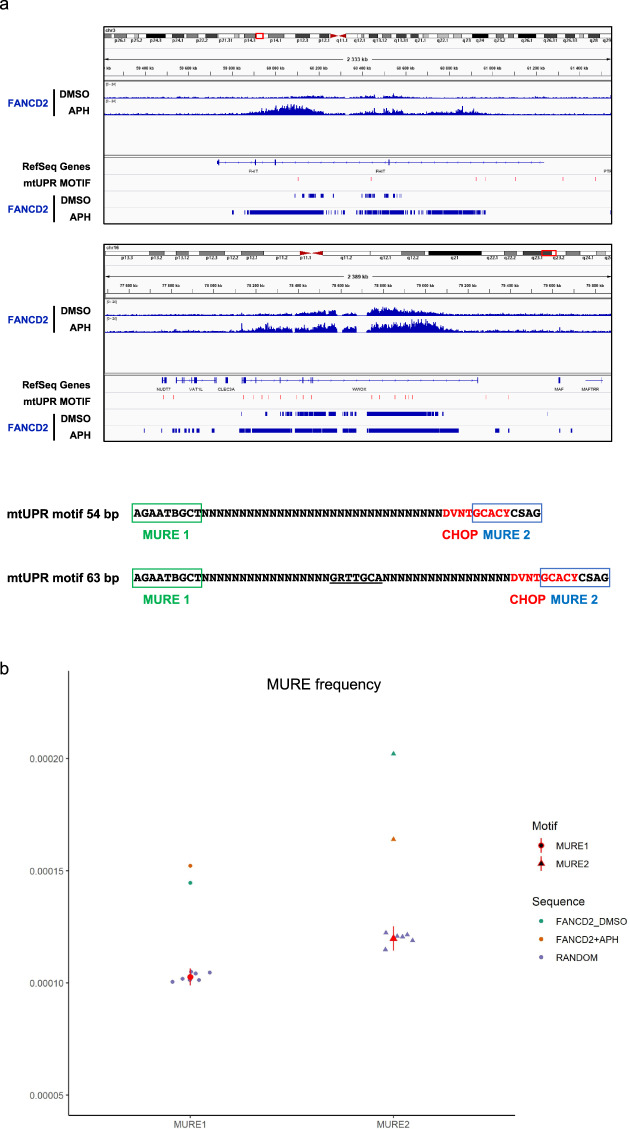
Fig. 3. Genome-wide analysis of FANCD2 targets identifies an enrichment for mitochondrial UPR-response elements at FANCD2-binding sites.
a IGV visualization of FANCD2 enrichment along the CFS genes FHIT and WWOX in the presence or absence of APH and the relative position of mitochondrial UPR (mtUPR) motifs (red tags) and FANCD2 peaks. FANCD2 ChIP seq data were scanned with regulatory sequence analysis tools (RSAT)-matrix-scan89,90 to identify instances of MURE1, MURE2 (mitochondrial UPR response elements) and CHOP motifs as they were defined by Aldridge et al. (2007) and Munch and Harper (2016)46,47. mtUPR motifs represent sequences of 54 or 63 bp with MURE1–CHOP–MURE2 consensus elements as indicated. The conserved CHOP consensus between MURE1 and MURE2 elements as reported by Aldridge et al. is underlined. Notice that a 10 bp consensus sequence for CHOP described in Munch and Harper is partially overlapping the MURE2 element. b Frequency of MURE1 and MURE2 elements at FANCD2-binding sites relative to a random control. The matches scored for each motif were compared to those detected in n = 7 (MURE1) or n = 6 (MURE2) independent sets of random sequences of identical length from human genome GRCh37-hg19. p values of the probability to obtain a similar score were calculated with the chi-square test. ****p = 5.84e−47 (MURE1), ****p = 1.91e−174 (MURE2), and ****p = 1.85e−69 (MURE1), ****p = 9.30e−70 (MURE2) for FANCD2 in mock (DMSO) and APH-treated conditions, respectively.