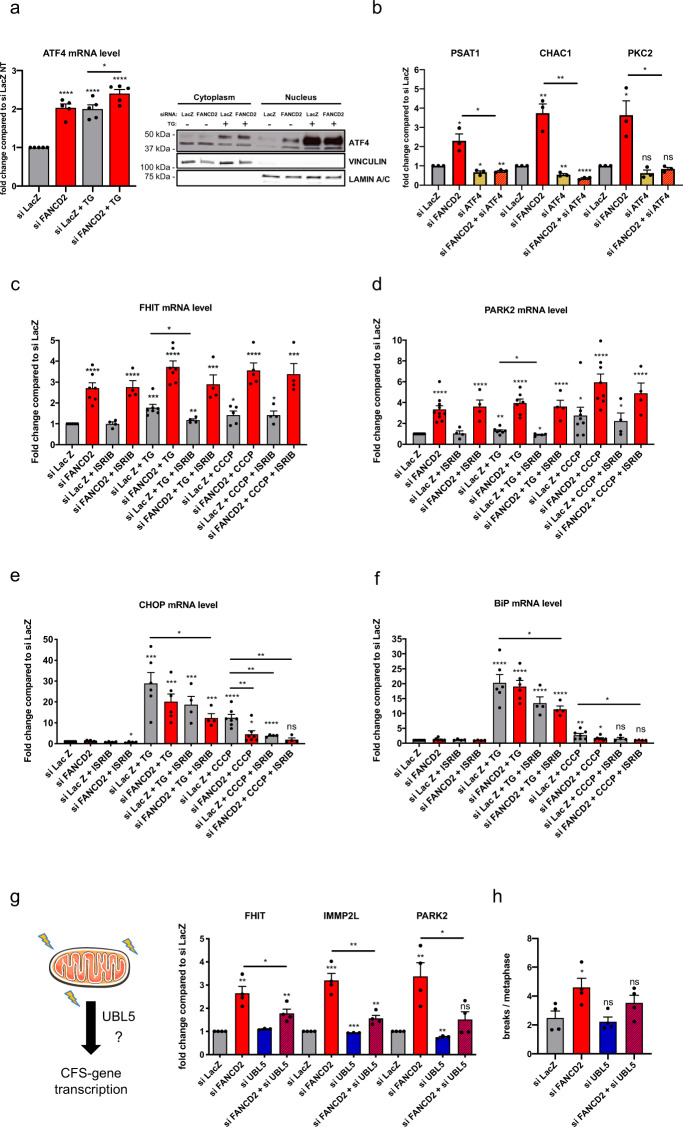
Fig. 6. FANCD2 functionally interacts with UBL5 in mitochondrial UPR signaling and CFS stability.
a ATF4 expression measured by RT-qPCR in siLacZ- and siFANCD2-transfected cells, treated or not with 1 µM TG (left panel). n = 5 independent experiments. ****p < 0.0001. Western Blot detection of ATF4 in cytoplasmic and nuclear fractions of siLacZ- or siFANCD2-transfected cells treated (+) or not (−) with 1 µM TG (right panel). Vinculin and lamin A/C were used as loading controls for cytoplasmic and nuclear fractions, respectively. b Expression of ATF4 target genes measured by RT-qPCR after control, FANCD2, ATF4, or FANCD2 and ATF4 siRNA transfection. n = 3 independent experiments. *p = 0.0194 (PSAT1_siFANCD2), *p = 0.0134 (PSAT1_siATF4), **p = 0.0015 (PSAT1_siFANCD2 + siATF4), *p = 0.0105 (PSAT1_siFANCD2 + siATF4 vs. PSAT1_siFANCD2); **p = 0.0048 (CHAC1_siFANCD2), **p = 0.015 (CHAC1_siATF4), ****p < 0.0001 (CHAC1_siFANCD2 + siATF4), **p = 0.0022 (CHAC1_siFANCD2 + siATF4 vs. CHAC1_siFANCD2); *p = 0.0239 (PKC2_siFANCD2), *p = 0.0201 (PKC2_siFANCD2 + siATF4 vs. PKC2_siFANCD2). c FHIT mRNA level measured by RT-qPCR in siLacZ- or siFANCD2-transfected cells treated or not with 1 µM TG or 10 µM CCCP and ISRIB (500 nM) for 8 h. n = 7 (siLacZ, siFANCD2, siLacZ + TG, siFANCD2 + TG), n = 5 (siLacZ + CCCP, siFANCD2 + CCCP), n = 4 (siLacZ + ISRIB, siFANCD2 + ISRIB, siLacZ + TG + ISRIB, siFANCD2 + TG + ISRIB, siLacZ + CCCP + ISRIB, siFANCD2 + CCCP + ISRIB) independent experiments. ****p < 0.0001 (siFANCD2), ****p < 0.0001 (siFANCD2 + ISRIB), ***p = 0.0004 (siLacZ + TG), ****p < 0.0001 (siFANCD2 + TG), **p = 0.0047 (siLacZ + TG + ISRIB), *p = 0.0241 (siLacZ + TG + ISRIB vs. siLacZ + TG), ***p = 0.0003 (siFANCD2 + TG + ISRIB), *p = 0.026 (siLacZ + CCCP), ****p < 0.0001 (siFANCD2 + CCCP), *p = 0.0202 (siLacZ + CCCP + ISRIB), ***p = 0.0001 (siFANCD2 + CCCP + ISRIB). d PARK2 mRNA level measured by RT-qPCR in siLacZ-transfected or siFANCD2-transfected cells treated or not with 1 µM TG or 10 µM CCCP and ISRIB (500 nM) for 8 h. n = 10 (siLacZ, siFANCD2), n = 8 (siLacZ + CCCP, siFANCD2 +CCCP), n = 7 (siLacZ + TG, siFANCD2 + TG), n = 4 (siLacZ + ISRIB, siFANCD2 + ISRIB, siLacZ + TG + ISRIB, siFANCD2 + TG + ISRIB, siLacZ + CCCP + ISRIB, siFANCD2 + CCCP + ISRIB) independent experiments. ****p < 0.0001 (siFANCD2), ****p < 0.0001 (siFANCD2 + ISRIB), **p = 0.0045 (siLacZ + TG), ****p < 0.0001 (siFANCD2 + TG), *p = 0.0312 (siLacZ + TG + ISRIB), *p = 0.0328 (siLacZ + TG + ISRIB vs. siLacZ + TG), ****p < 0.0001 (siFANCD2 + TG + ISRIB), *p = 0.022 (siLacZ + CCCP), ****p < 0.0001 (siFANCD2 + CCCP), *p = 0.0203 (siLacZ + CCCP + ISRIB), ****p < 0.0001 (siFANCD2 + CCCP + ISRIB). e CHOP mRNA level measured by RT-qPCR in siLacZ-transfected or siFANCD2-transfected cells treated or not with 1 µM TG or 10 µM CCCP and ISRIB (500 nM) for 8 h. n = 7 (siLacZ, siFANCD2, siLacZ + CCCP, siFANCD2 + CCCP), n = 6 (siLacZ + TG, siFANCD2 + TG), n = 4 (siLacZ + ISRIB, siFANCD2 + ISRIB, siLacZ + TG + ISRIB, siFANCD2 + TG + ISRIB, siLacZ + CCCP + ISRIB, siFANCD2 + CCCP + ISRIB) independent experiments. *p = 0.0221 (siFANCD2 + ISRIB), ***p = 0.0001 (siLacZ + TG), ***p = 0.0001 (siFANCD2 + TG), ***p = 0.0002 (siLacZ + TG + ISRIB), ****p < 0.0001 (siFANCD2 + TG + ISRIB), *p = 0.0382 (siFANCD2 + TG + ISRIB vs. siLacZ + TG), ****p < 0.0001 (siLacZ + CCCP), *p = 0.0477 (siFANCD2 + CCCP), ****p < 0.0001 (siLacZ + CCCP + ISRIB), **p = 0.0046 (siFANCD2 + CCCP vs. siLacZ + CCCP), **p = 0.0033 (siLacZ + CCCP + ISRIB vs. siLacZ + CCCP), **p = 0.0033 (siFANCD2 + CCCP + ISRIB vs. siLacZ + CCCP). f BIP mRNA level measured by RT-qPCR in siLacZ-transfected or siFANCD2-transfected cells treated or not with 1 µM TG or 10 µM CCCP and ISRIB (500 nM) for 8 h. n = 7 (siLacZ, siFANCD2, siLacZ + CCCP, siFANCD2 + CCCP), n = 6 (siLacZ + TG, siFANCD2 + TG), n = 4 (siLacZ + ISRIB, siFANCD2 + ISRIB, siLacZ + TG + ISRIB, siFANCD2 + TG + ISRIB, siLacZ + CCCP + ISRIB, siFANCD2 + CCCP + ISRIB) independent experiments. ****p < 0.0001 (siLacZ + TG), ****p < 0.0001 (siFANCD2 + TG), ****p < 0.0001 (siLacZ + TG + ISRIB), ****p < 0.0001 (siFANCD2 + TG + ISRIB), **p = 0.008 (siLacZ + CCCP), *p = 0.0244 (siFANCD2 + CCCP), *p = 0.0361 (siFANCD2 + TG + ISRIB vs. siLacZ + TG), *p = 0.0467 (siFANCD2 + CCCP + ISRIB vs. siLacZ + CCCP). g Left, graphical representation of mitochondrial UPR activation of CFS genes. Right, expression of large CFS genes measured by RT-qPCR after control, FANCD2, UBL5, or FANCD2 and UBL5 siRNA transfection. n = 4 (siLacZ, siFANCD2, siFANCD2 + siUBL5), n = 3 (siUBL5) independent experiments. **p = 0.014 (FHIT_siFANCD2), **p = 0.0062 (FHIT_siFANCD2 + siUBL5), *p = 0.0461 (FHIT_siFANCD2 + siUBL5 vs. FHIT_siFANCD2); ***p = 0.0003 (IMMP2L_siFANCD2), ****p < 0.0001 (IMMP2L_siUBL5), **p = 0.0062 (IMMP2L_siFANCD2 + siUBL5), **p = 0.0025 (IMMP2L_siFANCD2 + siUBL5 vs. IMMP2L_siFANCD2); **p = 0.0067 (PARK2_siFANCD2), ***p = 0.0004 (PARK2_siUBL5), *p = 0.0328 (PARK2_siFANCD2 + siUBL5 vs. PARK2_siFANCD2). h Chromosome fragility in cells transfected with control, FANCD2, UBL5, or FANCD2 and UBL5 siRNA and treated with 0.3 µM APH, scored as the mean number of breaks per metaphase. A total of 202 (siLacZ), 202 (siFANCD2), 194 (siUBL5), and 202 (siFANCD2 + siUBL5) metaphases were analyzed from n = 4 independent experiments. *p = 0.035 (siFANCD2). Error bars are standard error of the mean (SEM).