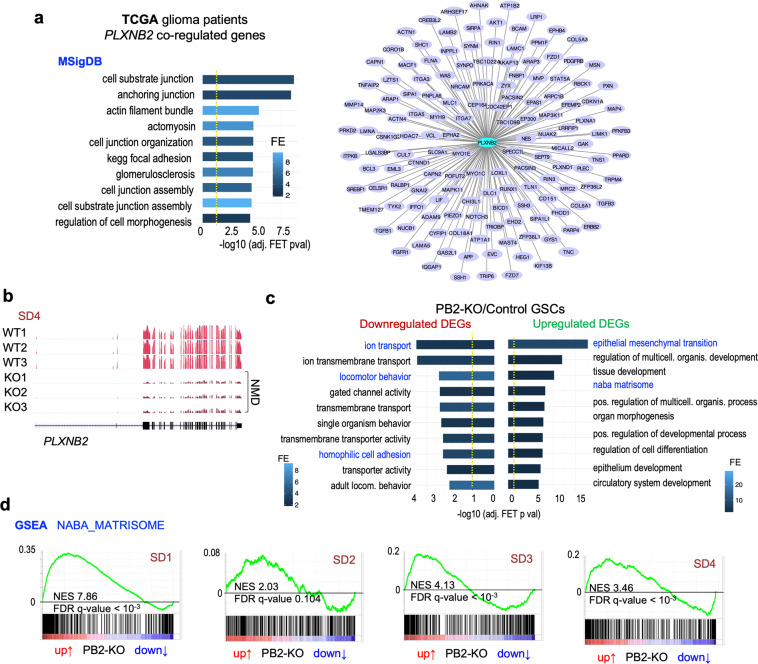
Fig. 6. Gene expression analyses link Plexin-B2 to biomechanical gene signatures in GBM.
a Genes that are correlated with PLXNB2 expression levels in GBM patients (Spearman correlation FDR < 10%, TCGA database) are enriched for pathways concerning cell adhesion, cell-substrate junctions, and actomyosin. Left, top 10 significant functional terms (MSigDB) are shown. Right, diagram of connectivity of coregulated genes with PLXNB2 in human GBMs, filtered for genes involved in cell adhesion, motility/invasion, and EMT. b Examples of RNA-seq read tracks for PLXNB2 mRNA in three GSC clonal lines of wild-type and PB2-KO genotypes from SD4 GSCs. Frameshift mutations in the PB2-KO lines lead to the reduction of PLXNB2 mRNA levels by nonsense-mediated decay (NMD). c Pathway enrichment analyses of up- and downregulated DEGs in response to Plexin-B2 KO (union of all DEGs from four GSC lines; FDR < 20%). Top ten significantly enriched terms (MSigDB) are shown. Dashed yellow lines denote −log10 (adjusted P value) = 0.05. d GSEA of expression changes in PB2-KO vs. control GSCs shows enrichment of Matrisome gene set (Naba et al.39) for all four GSC lines. The sinusoidal shapes of enrichment scores (green line) indicate enrichment of both up- and downregulated genes.