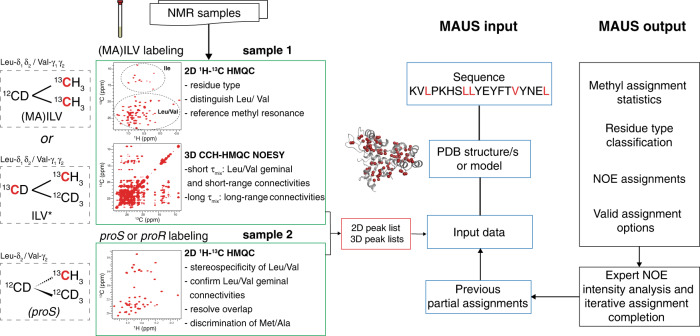
Fig. 2. Sample preparation, data collection, and methyl assignment workflow.
Two separate protein samples are recommended for generating the standard inputs of MAUS. Sample 1 uses non-stereospecific methyl labeling with either (i) 13C/1H labeling only at the methyls of Met, Ala, Leu, Val, and Ile residues, on a 12C/2H background or (ii) 13C/1H labeling at the methyls and 13C/2H at the sidechains of methyl-bearing residues on a 12C/2H background, creating linearized spin systems43, which can be used to unambiguously distinguish Leu/Val methyl resonances18 (Supplementary Figs. 4–6). When sample i is used, resonances of Leu/Val peaks can be partially distinguished using an automated chemical shift-based classifier within MAUS (“Methods”). Both samples can be used to define reference methyl chemical shifts using real-time (sample i) or constant-time (sample ii) 2D methyl-HMQC experiments. Long-range (up to 10 Å) NOEs are recorded using a 3D CM-CMHM SOFAST NOESY experiment (300 ms mixing time). A complementary NOESY experiment recorded with a short (typically 50 ms) mixing time is used to identify short-range NOEs, including between the geminal methyl resonances of Leu and Val residues. An additional protein sample 2 is prepared with stereospecific labeling (proS or proR) of Leu and Val methyls, which resolves spectral overlap in the 2D reference HMQC spectrum, and, together with sample 1, allows unambiguous determination of geminal methyl pairs for Leu/Val. 13C nuclei are displayed in red. Steps of NMR data analysis are colored green, with the resulting data set illustrated as a red rectangle. The standard input files for MAUS (blue) are the primary sequence, the PDB coordinate file or model structure, the 2D 1H-13C HMQC peak list, and two 3D CM-CMHM SOFAST NOESY peak lists (recorded with short and long mixing time). MAUS also accepts partial assignments that could be included in the user-annotated 2D 1H-13C HMQC list, together with a specification of allowed residue types, stereospecificity, and geminal methyl connectivities (if present) for each 2D reference peak. The output includes methyl assignment statistics, residue type classification, NOE assignments, and final lists of assignment options. Although MAUS does not consider the NOE peak intensities, this information can be evaluated by the user toward further reducing assignment ambiguities in the output lists.