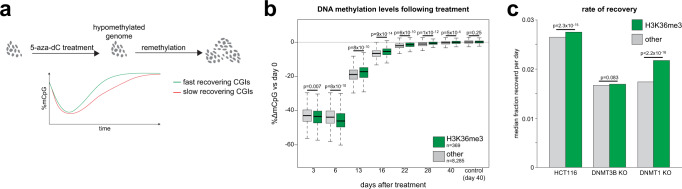
Fig. 4. H3K36me3 marked CGIs preferentially recover methylation following pharmacological hypomethylation.
a Schematic of experimental approach. 5-aza-dC is used to hypomethylate cells and the kinetics of methylation recovery are compared for different CGIs. b H3K36me3 marked CGIs preferentially recover methylation following 5-aza-dC treatment. Boxplots of relative methylation at H3K36me3 marked CGIs (n = 369) and all other CGIs methylated (n = 8285) in HCT116 cells. P-values are from two-sided Wilcoxon rank sum tests. Lines = median; box = 25th–75th percentile; whiskers = 1.5 × interquartile range from box. c The rate of methylation gain is higher at H3K36me3 CGIs in HCT116 cells. Barplot of the median normalised rate of methylation gain for H3K36me3 marked CGIs and other CGIs methylated in HCT116 cells and DNMT KO cells (n = 369, 371 and 284 for K36me3 CGIs and 8285, 8284 and 6519 for other CGIs in HCT116, DNM3B KO and DNMT1 KO, respectively). The rate of methylation recovery was estimated by fitting linear models to the RRBS data for each CGI. P-values are from two-sided Wilcoxon rank sum tests. Source data are provided as a Source Data file.