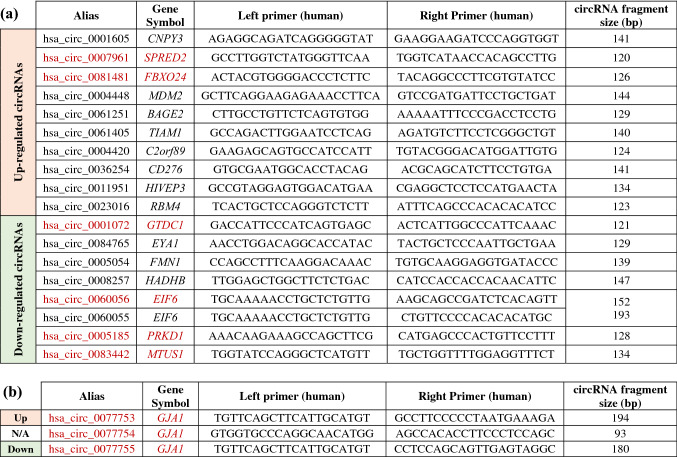
Table 1.
Selection of 18 circRNAs involved in cancer-related and/or epithelial polarity pathways for RT-qPCR validation.
(a) Ten up-regulated (of 75 significantly up-regulated) and eight down-regulated (of 46 significantly down-regulated) circRNAs in Cx43-KO-S1 as compared to S1 cells were chosen for further validation based on whether the genes from which they originate were involved in cancer-related and/or epithelial polarity pathways. (b) The three isoforms of circRNAs that originate from Cx43 (GJA1) mRNAs were also chosen due to their relevance to the 3D risk-progression culture model. One Cx43 circRNA isoform, hsa_circ_0077754, was not detected by the microarray due to its short size of 93 bp, since library size-selections usually excludes circRNAs < 200 nt long58. Alias represents the circRNAs probe name as annotated by CircBAse59. Gene Symbol denotes the gene of the transcript from which this circRNAs originate. Primers were designed using CircularRNA Interactome in silico tool60 and purchased from Eurofins (Canada). The circRNA fragment size represents the size of the resulting amplified PCR fragment. The circRNAs highlighted in red font were significantly dysregulated in Cx43-KO-S1 as compared to S1 cells in 3D, as validated through RT-qPCR (shown in Fig. 4).