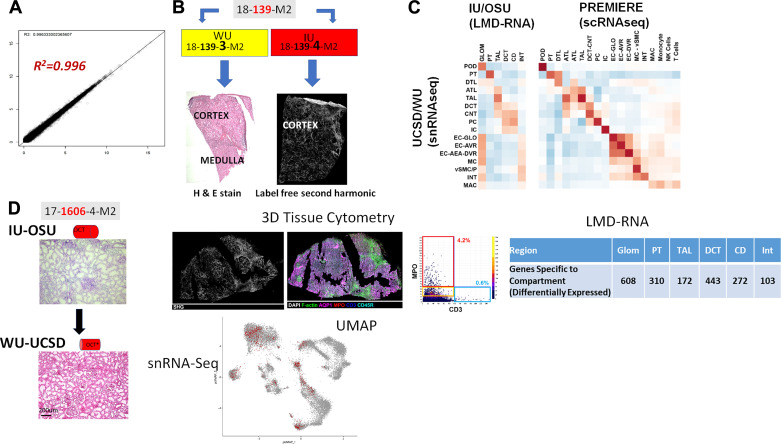
Figure 5.
Testing the “follow the tissue” pipeline with data generated from pilot samples and demonstrating feasibility of multiple technologies on limited tissue sample across sites. A: high correlation of bulk RNA-seq from adjacent samples shipped and processed at different sites. B: adjacent samples from same patient shipped to different tissue investigative sites (TISs) show different types of information from different technologies in Kidney Precision Medicine Project (KPMP) with histological tissue composition verification for snRNAseq at WU-UCSD and label-free second harmonic generation highlighting extent of fibrosis at IU-OSU. C: correlation between cell types and regions among snRNA-seq, laser microdissection (LMD)transcriptomics of the indicated regions by IU/OSU, and between snRNA-seq and scRNA-seq. D: economizing tissue usage by rotating fresh optimal cutting temperature (OCT) frozen kidney tissue between IU and WU/UCSD. Here. frozen sections were processed and analyzed for 3-D tissue cytometry and LMD-RNA analysis, and then the block was shipped to WU/UCSD for histological registration and subsequently generating snRNA-seq. Genes specific to each LMD compartment were determined by their upregulation in that compartment compared with other compartments as well as differential expression at a nominal P < 0.001. The UMAP plot shows the sample (∼900 nuclei) contributing to many of the cortical cell types in the kidney snRNAseq atlas (∼18,000 nuclei) (11). IU, Indiana University; OSU, Ohio State University; UCSD, University of California, San Diego; WU, Washington University in St. Louis; UM, University of Michigan; UCSF.