Abstract
JC virus encodes an important regulatory protein, known as Agnoprotein (Agno). We have recently reported Agno’s first protein-interactome with its cellular partners revealing that it targets various cellular networks and organelles, including mitochondria. Here, we report further characterization of the functional consequences of its mitochondrial targeting and demonstrated its co-localization with the mitochondrial networks and with the mitochondrial outer membrane. The mitochondrial targeting sequence (MTS) of Agno and its dimerization domain together play major roles in this targeting. Data also showed alterations in various mitochondrial functions in Agno-positive cells; including a significant reduction in mitochondrial membrane potential, respiration rates and ATP production. In contrast, a substantial increase in ROS production and Ca2+ uptake by the mitochondria were also observed. Finally, findings also revealed a significant decrease in viral replication when Agno MTS was deleted, highlighting a role for MTS in the function of Agno during the viral life cycle.
Keywords: Mitochondria, JC virus, agnoprotein, progressive multifocal leukoencephalopathy, neurological diseases, polyomavirus, SV40, BKV, TOM, TIM
1. Introduction
Viruses are obligatory intracellular microorganisms and employ various strategies to modify the host-cell environment by targeting the host-protein complexes, network signaling pathways, organelles and thereby create a more conducive environment in order to survive and successfully complete their life cycle (Brito and Pinney, 2017; Franzosa and Xia, 2011). One of the organelles targeted by the viruses is mitochondria, the energy powerhouse of the cells (Boya et al., 2004). In addition to producing ATP as an energy source (Boyman et al., 2019), mitochondria are also involved in various cellular mechanisms, including apoptosis (Boya et al., 2003; Thomson, 2001), antiviral immunity (Cloonan and Choi, 2012; Foy et al., 2005; Li et al., 2005) and Ca2+ homeostasis (Li et al., 2007). It is a double membraned organelle, harboring its own circular DNA genome and translation machinery. Mitochondrial genome encodes 13 mitochondria proteins, which are mostly subunits of respiratory chain complexes, I, III, IV and V; 2 rRNAs and 22 mitochondrial t-RNAs (Anand and Tikoo, 2013). In addition to its own encoded proteins, mitochondria import the majority of its structural and functional proteins encoded by nuclear DNA (Anand and Tikoo, 2013). These imported proteins have either amino terminal or internal mitochondrial targeting sequence (MTS) (Pfanner, 2000), the length of which varies depending on the protein type (10-80 aa long). MTS contains predominantly positively charged amino acid sequences. Once translocated into the mitochondrial matrix, MTS sequences are cleaved and removed inside the matrix; and matured proteins are then distributed into the various locations in the mitochondria depending on their functional properties (Pfanner, 2000; Wiedemann and Pfanner, 2017). A number of reports indicate that various viruses target mitochondria through their own-encoded proteins to modulate the different aspect of mitochondrial functions in order to maximally utilize the host environment. These modulated functions include the Ca2+ homeostasis, mitochondrial membrane potential, mitochondrial stress due to the production of excessive reactive oxygen species (ROS) and mitochondrial antiviral immunity (Anand and Tikoo, 2013; Boya et al., 2004).
JC virus (JCV) is a neurotropic human polyomavirus and contains a small double-stranded DNA genome (~ 5kb). It asymptomatically infects individuals during childhood and establishes a latent infection in several tissues and organs, including, kidneys, peripheral blood B-lymphocytes, tonsillar stromal cells and even in the brain (Monaco et al., 1998; White et al., 1992). Under severe immunosuppressive conditions, JCV reactivates itself from latency and initiates a primary infection in oligodendrocytes and astrocytes in the central nervous system (CNS); and causes a fatal demyelinating disease known as progressive multifocal leukoencephalopathy (PML) (Ferenczy et al., 2012; Saribas et al., 2018a). PML develops in a subpopulation of immunocompromised patients including those with AIDS, cancer and multiple sclerosis (Berger, 1987, 2000; Ferenczy et al., 2012; Kleinschmidt-DeMasters and Tyler, 2005; Langer-Gould et al., 2005; Padgett, 1971; Van Assche et al., 2005). JCV was also reported to infect neurons (Dang et al., 2012; Du Pasquier et al., 2003; Soleimani-Meigooni et al., 2017; Wuthrich et al., 2016; Wuthrich et al., 2009), and cause another JCV-induced brain pathology called “granule cell neuronopathy” (GCN) (Dang et al., 2012; Du Pasquier et al., 2003; Soleimani-Meigooni et al., 2017; Wuthrich et al., 2016; Wuthrich et al., 2009). Recent reports also indicate that the extracellular vesicles produced by choroid plexus epithelial and the meningeal cells in the CNS may play roles in the dissemination of infectious JCV particles in the brain (Morris-Love et al., 2019; O’Hara et al., 2018; O’Hara et al., 2020).
JCV encodes a limited number of regulatory and structural proteins from its early and late coding regions including large T antigen, small t antigen, T’-proteins, Agnoprotein (Agno), ORF1, ORF2, VP1, VP2 and VP3 (Frisque et al., 1984; Saribas et al., 2018a). Among those, Agno, encoded by the JCV late coding region, was shown to play critical roles during the viral life cycle (Saribas et al., 2016; Sariyer et al., 2011). In the absence of its expression, JCV is unable to sustain its productive life cycle (Sariyer et al., 2011). Agno is a small and highly basic phospho-protein (71 aa long) and primarily localizes to the cytoplasmic compartment of the infected cells with high concentrations accumulating at the perinuclear area (Ellis and Koralnik, 2015; Gerits and Moens, 2012; Nomura et al., 1983; Rinaldo and Hirsch, 2007; Rinaldo et al., 1998; Saribas et al., 2016; Saribas et al., 2012). However, a small portion of the protein is also consistently detected in the nucleus (Okada et al., 2001; Saribas et al., 2012), suggesting regulatory roles for Agno at both compartments of the infected cells. One of the remarkable features of this protein is its ability to form highly stable homodimers and oligomers mediated by its major α-helical domain (Saribas et al., 2013; Sariyer et al., 2011). Another interesting feature of Agno is its release from the Agno-positive cells into the extracellular space (Otlu et al., 2014; Saribas et al., 2018a) and how its major α-helical domain plays a significant role in this process (Otlu et al., 2014; Saribas et al., 2018a). The function of the released Agno is yet to be determined, however, initial characterization studies showed that it strongly interacts with unidentified cell surface components (Saribas et al., 2018b) and is perhaps subsequently taken up by the cells (Craigie et al., 2018), suggesting that it exerts its activity either through the cell surface or through entering the cells. Additionally, Agno was implicated to be involved in various aspects of the JCV life cycle, including viral transcription and replication (Safak et al., 2001), virion formation (Sariyer et al., 2006; Suzuki et al., 2012), functioning as a viroporin (Suzuki et al., 2013; Suzuki et al., 2010), deregulation of cell cycle progression (Darbinyan et al., 2002) and targeting lipid droplets (Unterstab et al., 2010) and evasion of the host immune system (Manzetti et al., 2020).
Moreover, we, in collaboration with others, recently resolved the three dimensional (3D) structure of Agno (Coric et al., 2017; Coric et al., 2014) revealing that it contains two α-helical domains designated as minor and major α-helices, which span from Leu6 to Lys13 and from Arg24 to Phe39 respectively and the rest of the protein adopts an intrinsically disordered conformation (Coric et al., 2017). Such disordered regions are known to provide considerable flexibility to any protein to diversify its binding partners and thereby amplify their functions in cells (Dyson and Wright, 2005; Fink, 2005; Ou et al., 2012). It is conceivable that the disordered regions within Agno also play similar functions. Furthermore, our recent proteomic studies revealed that Agno primarily interacts with the cellular proteins containing “coiled coil” motifs, and targets numerous protein networks such as tRNA synthases, translation initiation factors, ubiquitin-proteasome complexes; and organelles including nucleus, endoplasmic reticulum, Golgi apparatus and mitochondria (Saribas et al., 2020). With respect to its mitochondrial targeting, Agno shows interactions with the mitochondrial proteins involved in the structural component of the mitochondrial protein import pathways, electron transport chain (ETC) complexes and metabolic enzymes (Saribas et al., 2020).
Agno harbors several functional domains, including a bipartite nuclear localization signal domain (NLS) and the overlapping dimerization and nuclear export signal (NES) domains (Saribas et al., 2019). Additionally, our recent NMR and proteomic studies as well as the previous bioinformatics predications indicated that the N-terminus of Agno contains a putative mitochondrial targeting sequence (MTS, aa 1-14, MVLRQLSRKASVKV), which can facilitate its translocation into mitochondria, thereby modulating the mitochondrial functions. Thus, in this study, we further characterized the functional consequences of mitochondrial targeting by Agno and showed its co-localization with the mitochondrial network and co-fractionation with mitochondrial outer membrane (MOM) fractions. Our data also showed a significant decrease in various mitochondrial functions such as mitochondrial membrane potential, respiration and ATP production. Conversely, a substantial increase in both ROS production and Ca2+ uptake by mitochondria was also observed in Agno-positive cells. We also observed a significant reduction in the replication efficiency of JCV when the Agno MTS is deleted, highlighting the importance of interplay between Agno and mitochondria during the viral replication cycle.
2. Materials and methods
2.1. Cell lines
SVG-A is a human cell line established by transforming the primary human fetal glial cells with an origin-defective SV40 mutant (Major et al., 1985). SVG-A cells provide detailed microscopic images due to their relatively large cell size and support JC virus replication. HEK293 cells (ATCC, catalog no. CRL-326) are transformed human embryonic kidney cells and can be transfected with calcium-phosphate method with high efficiency. These have a relatively small cell size and therefore do not provide detailed microscopic images. HEK393T cells do not support JC virus replication. All cell lines were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) (Life Technologies, catalog no. 31600–034) supplemented with 10% heat inactivated fetal bovine serum (FBS) (Gemini, catalog no. 100-106) and antibiotics [penicillin–streptomycin 100 μg/ml (Gemini, 400-109), ciprofloxacin 10 μg/ml (Sigma catalog no. 17850) and tetracycline (ThermoFisher, catalog no. A39246, 1μg/ml)]; and maintained at 37°C in a humidified atmosphere with 7% CO2. Primary human fetal astrocytes (PHFA) were obtained from the Temple University Comprehensive NeuroAIDS Center, fed with mixed glial growth media (DME:F12 media supplemented with glutamax (ThermoFisher, cat. number: 35050061), insulin (Sigma, catalog no. I0516-5ml, 10 μg/ml final), 10 % FBS, L-glutamine (Corning, catalog no. 25-005 Cl) and gentamycin (Life Technologies, catalog no. 15750-060); and maintained at 37°C in a humidified atmosphere with 7 % CO2.
2.2. Creation of the expression plasmids and viruses
Following plasmid constructs were used in this study: the construction of the pCGT7-JCV Agno WT (1-71) plasmid was previously reported (Otlu et al., 2014; Saribas et al., 2018b) and pCGT7-JCV Agno (15-71) mutant was generated by deleting 1-14 aa from Agno wild-type. Adenovirus (Ad)-Agno expression system was created as follows: Agno WT (1-71) and Agno Mutant (15-71) DNA sequences were separately subcloned into pDC515(IQ) adenovirus vector (https://microbix.com) at EcoRI/SalI sites by PCR-based cloning methodology. Subsequently, each plasmid was co-transfected with another shuttle plasmid (pBHG frt (del) E1 E3 FLP) into 293-IQ (https://microbix.com) to create a recombinant Adenovirus expression system for Agno WT and Agno mutant (15-71). Ad-Agno recombinant viruses were then selected, propagated and used in various experiments. Bluescript KS(+) –JCV Mad-1 Agno (15-71) virus was created by deleting the aa 1-14 from the viral background and introducing a Met (ATG) at the 15th aa of Agnoprotein using QuikChange mutagenesis (Agilent, cat. number: 200521). Bluescript KS(+)-JCV Mad-1 WT was created by inserting the JCV Mad-1 sequence (GenBank accession #: J02226.1). In addition, Agno WT (1-71) and its various deletion mutants were fused to a green fluorescent protein (GFP) by cloning them at BamHI/EcoRI sites of pEGFP-N1 vector and the resulting plasmids designated as follows: pEGFP-N1-Agno WT (1-71), pEGFP-N1-Agno mutant (1-14), and pEGFP-N1-Agno mutant (15-71). The creation of the pCGT7-ORF2 plasmid was previously reported (Saribas et al., 2018a).
2.3. Mitochondria isolation
The tissue culture plates (100 mm, Becton Dickenson, catalog no. 353003), were first treated with Poly-D-lysine hydrobromide (Sigma, catalog no. P6407-5MG) in order to have a firm cell adherence onto the surface of the plates. The HEK293T cells were then plated onto these plates in 80% confluency the day before transfections. Next day, cells were transfected with pCGT7-Agno WT expression plasmid (30 μg/plate) using the calcium phosphate precipitation method as described previously (Graham, 1973). At 36 h post-transfection, mitochondria were isolated from the untransfected (control) and transfected cells using a “mitochondria isolation kit” (ThermoFisher, catalog no. 89874) following the manufacturer’s protocols. Isolated mitochondria were then analyzed by Western blotting for detection of Agno using α-Agno (Del Valle et al., 2002) and α-COX2 (Abcam, catalog no. ab3298).
2.5. Affinity purification of mitochondria
Mitochondria were purified using an affinity-based purification system according to the manufacturer’s recommendations (Miltenyi Biotec, catalog no. 130-094-532). Briefly, HEK293 cells were plated onto 100 mm tissue culture dishes treated with Poly-D-lysine in 80% confluency the day before transfections. On the next day, cells were transfected with either pCGT7-Agno WT, pCGT7-Agno (15-71) mutant or pCGT7-ORF2 (Saribas et al., 2018a) expression plasmids, (30 μg DNA/plate) using the calcium phosphate precipitation method as described previously (Graham, 1973). At 36h posttransfection, cells were harvested by trypsin treatment, washed with 1xPBS by centrifugation (400g) and resuspended in 1 ml ice-cold lysis buffer (Miltenyi Biotec, catalog no. 130-094-532) (1x107 cells/sample) supplemented with protease inhibitors (Sigma, catalog no. P8340). Cells were homogenized using a 28-gauge needle (BD insulin syringe, catalog no. 329424). The stroke numbers in homogenization were empirically determined prior to experiments using untransfected cells. The 1 ml homogenate was then transferred to a 15 ml conical tube, mixed with 9 ml ice-cold Separation Buffer (Miltenyi Biotec, catalog no. 130-094-532) and incubated with 50 μl of α-TOM22 microbeads (Miltenyi Biotec, catalog no. 130-094-532) to magnetically labeled mitochondria. After 3h incubation at 4 °C, antibody-homogenate mixture (10 ml) was loaded onto LS Columns (Miltenyi Biotec, catalog no. 130-094-532) to capture the labeled mitochondria by using a miniMACS separation unit (Miltenyi Biotec, catalog no. 130-042-102). After draining the unbound proteins and organelles by dripping, columns were washed with Separation Buffer (3 x 3 ml) by dripping. The columns were then placed into 15 ml conical tubes and the mitochondrial content was flushed out by applying 1.5 ml Separation Buffer, followed by firmly pushing the syringe plungers into the columns. Mitochondria were concentrated by centrifugation (13,000 g, for 5 min at 4 °C) and the mitochondrial pellet was resuspended in 200 μl in TNN lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1.0 % NP-40) by pipetting in and out and incubating them on a rocking platform at 4 °C for 30 min. Samples (40 μl each) were then separated on a 4-20% gradient SDS-PAGE (Bio-Rad, catalog no. 4561096) and analyzed by Western blotting using specific antibodies described under relevant figure legends.
2.4. Examining of the co-localization of Agno with mitochondria by immunocvtochemistry
SVG-A cells were plated onto 100 mm tissue culture dishes at 80% confluency the day before transfection. Next day, cells were transfected with pCGT7-Agno WT plasmid (8 μg/plate) using lipofectamine™ 3000 transfection reagent according to the Manufacturer’s recommendations (Life Technologies, catalog no. L-3000008). The lipofectamine/DNA ratio was maintained at the 2:1 ratio in the transfection mixture. At 24 h posttransfection, the transfected cells were transferred onto glass slide chambers (Nunc, catalog no. 154461) and incubated for an additional 24h. The cells were then washed with 1xPBS and fixed with 4% paraformaldehyde (Sigma, catalog no. 1004965000) prepared in 1xPBS supplemented with 0.05 % Triton X-100 (Sigma, catalog no. T8787) for 5 min, blocked with 5% bovine serum albumin (BSA) (Sigma, catalog no. A2058-25G) prepared in 1xPBS for 30 min. (Sigma, catalog no. 1004965000) for 30 min. The fixed cells were then probed with a combination of α-T7 (EMD Millipore, catalog no. 69522, monoclonal) (1:200 dilution) and α-COX4 (GeneTex, GTX101499) polyclonal (1:200 dilution) primary antibodies prepared in 1xTBST buffer (50 mM Tris-HCL, pH 7.4, 150 mM NaCl, 0.1 % Tween 20) overnight. Cells were washed three times with 1xTBST buffer for 5 min intervals and subsequently incubated with either a fluorescein isothiocyanate (FITC)-conjugated goat α-rabbit (Abcam, catalog no. Ab6717) or Rhodamine-conjugated goat α-mouse (Millipore, catalog no. AP124R) secondary antibodies for 45 min. Cells were then washed with 1xTBST buffer three times for 5 min each, incubated with DAPI for 5 min [ThermoFisher, 4′,6-Diamidino-2-Phenylindole, Dihydrochloride, catalog no. D1306) (300 ng/ml prepared in 1xPBS)] to stain the nucleus, mounted using “ProLong® Gold Antifade” mounting medium (Life Technologies, catalog no. P36934) and dried overnight under a cover slip (ThermoFisher, catalog no. 102450). Slides were then examined under a fluorescence microscope (Leica, DMI-6000B, objective: HCX PL APD 40x/1.25 oil, employing LAS AF operating software) for visualization of the proteins of interest.
In addition, the pEGFP-N1-Agno WT (1-71), pEGFP-N1-Agno mutant (1-14), and pEGFP-N1-Agno mutant (15-71) and pEGFP-N1 vector alone were separately transfected into the SVG-A cells plated onto glass slide chambers (Nunc, catalog no. 154461) using lipofectamine™ 3000 transfection as described above. At 24h posttransfection, the cells were fixed with 4% paraformaldehyde prepared in 1xPBS supplemented with 0.05 % Triton X-100 for 10 min and blocked with 5% BSA for 30 min. The fixed cells were then probed with primary α-COX4 (GeneTex, GTX101499) (1:200 dilution) overnight. Next day, cells were incubated with rhodamine-conjugated goat α-rabbit (Millipore, catalog no. AP132R) secondary antibody (1:200 dilution) for 45 min. After staining the nuclei of the cells with DAPI as described above, the slides were mounted and examined under a fluorescence microscope (Leica, DMI-6000B, objective: HCX PL APD 63x/1.25 oil, employing LAS AF operating software) for visualization of the proteins of interest (Supplement 2).
2.6. Mitochondria membrane fractionation
Tissue culture plates (100-mm culture dishes) were first coated with Poly-D-lysine, onto which HEK293T cells were plated at 80 % confluency the day before transfection. Cells were then transfected with either pCGT7 Agno WT or pCGT7 Agno (15-71) mutant expression plasmids (30 μg/plate) employing the calcium-phosphate transfection method. At 30 h post-transfection, cells were washed with cold 1xPBS, scraped off the culture plate on ice and spun at 600 g at 4°C for 5 min. Cells were washed again with 1xPBS plus 1% protein inhibitor cocktail (Sigma, catalog no. P8340) at 600 g at 4°C for 5 min. The cell pellet was suspended in mitochondria isolation buffer [320 mM Sucrose, (Sigma, catalog no. S0389), 1 mM EDTA (Sigma, catalog no. E6758), 10 mM Tris-HCl; pH 7.4 (Sigma, catalog no. 10812846001)] plus 1/200 protease inhibitor cocktail (Sigma, catalog no. P8340) and lysed by passing the samples through the 27-gauge syringe (BD catalog no. 309623) ten times. The homogenate was centrifuged at 600 g for 5 min to precipitate the cell debris at 4°C, and the supernatant was further centrifuged at 10,000 g for 10 min at 4°C to precipitate the mitochondrial fractions. The final supernatant was used as the cytosolic fraction. The isolated mitochondria pellet was then washed one more time with mitochondria isolation buffer followed by a centrifugation at 10000 g for 10 min 4°C. The pellet containing mitochondria was resuspended in mitochondria isolation buffer (100 μl) and treated with proteinase K (100 ug/ml) (Sigma, catalog no. P2308) for 5 min in the absence or presence of increasing concentration of digitonin (0.1, 0.25 and 0.5 % final concentration) (Sigma, catalog no. D141) for 15 min at 25°C. The reactions were stopped by adding the SDS-PAGE sample buffer and heating them at 95°C for 5 min. The Western blots were probed with α-Agno (polyclonal), α-COX4 (COX4 = mitochondria inner membrane protein) (polyclonal, GeneTex, catalog no. GTX101499) and α-VDAC1 (polyclonal, GeneTex, catalog no. GTX114187). Voltage-dependent anion channel 1 protein (VDAC1) is a mitochondria outer membrane protein.
2.7. Measurement of the ATP production levels in Agno-positive cells
ATP levels in cells were assessed using a ATP determination kit (Molecular Probes, catalog no. A22066) following the manufacturer’s protocols. This is a luciferase assay-based kit and the luminescence signal emitted by the cell extracts is directly proportional to the ATP content of cells. HEK293T cells were plated onto 60 mm tissue culture plates dishes (Life sciences, catalog no. 353002) treated with poly-D-lysine at ~70 % confluency in triplicates. They were transfected with either pCGT7-Agno WT (1-71), pCGT7-Agno (15-71) mutant, pCGT7-ORF2 (1-72) (Saribas et al., 2018a) (a negative control) or vector alone (pCGT7) (a negative control) (30 μg DNA/plate). At 24 h posttransfection, cells were lysed in lyses buffer (25 mM Tris-Cl, pH 7.8, 2 mM EDTA and 1% Triton-X 100) and the lysates were used in luciferase assay following manufacturer’s protocols. Chemoluminescence was measured by using Zylux FB12 single tube luminometer (Biocompare). The results were expressed as a “relative luciferase activity”.
2.8. Measuring the Ca2+ influx into mitochondria
SVG-A cells were grown on 60 mm tissue culture dishes at ~ 80 % confluency and transfected with either pCGT7-Agno WT or pCGT7 alone (control) (8 μg DNA/plate) using lipofectamine™ 3000 transfection reagent according to the manufacturer’s recommendations. Next day, the transfected cells were transferred onto the 35-mm glass bottom tissue culture plates (MatTek, catalog no. P35GC-14-C) and grown 16 h in the same tissue culture plates. Then, the culture media was removed and cells were simultaneously treated with Rhod-2/AM (2 μM final concentration, Life Technologies, catalog no. R1245MP) for 50 min and Fluo-4/AM (5 μM, Life Technologies, catalog no. F14201) for 30 min in cell medium as previously described (Madesh et al., 2005; Mallilankaraman et al., 2012a; Mallilankaraman et al., 2012b). Next, cell plates were washed twice with 1xPBS, replenished with fresh media, mounted in an open perfusion micro incubator (Harvard Apparatus, PDMI-2), maintained at 37°C and imaged using confocal microscope (510 Meta; Carl Zeiss Inc.). After 1 min of baseline recording, cells were treated with ionomycin (calcium ionophore) (Sigma, catalog no. I0634-1mg) and confocal images were obtained every 3 seconds (at 488- and 561- nm excitations) using a 40x oil objective during continuous live-cell imaging. Image analysis and quantitation of fluorescence intensities associated with individual cells was calculated using the ZEN 2010 software (https://www.zeiss.com/microscopv/us/products/microscope-software/zen-core.html).
2.9. Cell sorting by flow cytometry
The SVG-A cells were plated onto 100 mm tissue culture dishes at ~ 80 % confluency and were co-transfected with pEGFP-N1 (NovoPro Bioscience Inc, catalog no. V12021), and pCGT7-Agno WT in 1:6 ratio to achieve a high co-transfection efficiency for GFP and Agno (8 μg DNA total). At 24 h post-transfection, the transfected cells were washed twice with 1xPBS, trypsinized, which is neutralized with culture media (DMEM) containing 10% FBS, washed again twice with 1xPBS and finally resuspended in 1xPBS. The cells were then subjected to cell sorting based on the GFP expression at the “Flow Cytometry Core Facility” (Lewis Katz School of Medicine, Temple University) (https://medicine.temple.edu/research/facilities-and-services/flow-cytometry-core). The sorted GFP-positive cells were further examined to demonstrate the coexpression of Agno and GFP in majority of sorted cells by immunocytochemistry. More than 85% of the cells were observed to be co-transfected.
2.10. Viral infection/transduction
SVG-A and primary cells were transduced using adenoviral vectors at a defined multiplicity of infection (MOI) (10 PFU/cell). Briefly, 1 x 106 cells were plated in 6-well culture plate (Corning corporation, catalog no. 3506) and after 4 h incubation, the culture media was aspirated and cells were transduced using adenovirus (Ad) expression systems [Ad-Null, Ad-Agno WT, or Ad-Agno (15-71) mutant] prepared in 500 μl OptiMEM media (Lifescience Technologies, catalog no. 31985-070) as inoculum. Cells were incubated with this inoculum for 30 min at 37°C in a humidified atmosphere with 7 % CO2 with gentle shaking at every 5 min to allow the spread of inoculum over the cells. Following incubation, cells were fed with 4 ml of the complete DMEM and were further incubated overnight at 37°C in a humidified atmosphere with 7 % CO2. At 24 h post-transduction, the transduced cells were used for different assays.
2.11. Determining the oxygen consumption rates (OCR) by using Seahorse XFe96 Flux Analyzer
SVG-A and PHFA cells grown in 6-well tissue culture plates (Corning, catalog no. 07-200-83) were transduced with Ad-Null and Ad-Agno WT viruses. At 18 h post-transduction, the transduced cells were re-seeded (20,000 cells/plate) onto 96-well plates (Agilent, catalog no. 101085-004) in 200 μl of growth medium and incubated in a tissue culture incubator overnight at 37°C in a humidified atmosphere with 5 % CO2. One hour prior to assay, the culture media was replaced by XF assay media (180 μl (Agilent Technologies, catalog no. 102353-100) and the cells were incubated in a 37°C incubator without CO2 for 1 h to allow the system to pre-equilibrate with the XF assay medium [supplemented with 25 mM glucose (Sigma, catalog no. D8270), 2 mM glutamine (Sigma, catalog no. 49419-25G) and 1 mM sodium pyruvate (Sigma, catalog no. S8636; pH 7.4)]. Cells were then sequentially treated with oligomycin (1 uM) to block mitochondrial ATP-liked respiration, FCCP (3 μM) to quantify maximal respiration and rotenone plus antimycin A (1 μM) to completely inhibit all mitochondrial respiration. These inhibitors were supplied with assays kit. The OCR measurements of basal respiration (base OCR – non-mito respiration (post-Rot/AA), ATP-coupled respiration (post-oligo OCR–base OCR), Max respiratory capacity (post-FCCP OCR–post-Rot/AA), Spare respiratory capacity (post-FCCP OCR–basal OCR) and Proton leak (post-Oligo OCR–post-Rot/AA OCR) were performed using the Seahorse Bioscience Extracellular Flux Analyzer XF96e (Seahorse Bioscience) and the XF Cell mito stress kit reagents (Agilent Technologies, catalog no. 103015-100) following the manufacturer’s recommendations. The results were then normalized according to cell-protein concentration.
2.12. Tetramethylrhodamine-6-maleimide (TMRM) assay to determine the mitochondrial membrane potential
SVG-A cells were grown in 60-mm tissue culture plates and co-transfected with expression plasmids in the following combination: pGFP-N1 plus pCGT7-Agno WT or pGFP-N1 plus pCGT7-Agno (15-71) mutant (8 μg DNA total) in 1:6 ratio using Lipofectamine™ 3000 reagent. At 18 h post-transfection, cells were trypsinized, washed with 1xPBS, resuspended in 1xPBS and subjected to flow cytometry to sort GFP-positive cells as described above. These cells were then plated on 35-mm glass bottom poly-L-lysine coated plates (MatTek, catalog no. P35GC-0-14-C) for 5 hours to adhere them on the well surface and were then treated with TMRM (Molecular Probes kit, catalog no. T668) following the manufacturer’s recommendations. In brief, culture medium was removed and cells were treated with DMEM containing 5 uM of TMRM and counterstained with Hoechst 33342 (5 μg/ml) (Molecular Probes, catalog no. H1399) and incubated in tissue culture incubator at 37°C for 15 min. After staining, cells were washed twice with 1xPBS and live cells were imaged by using a Carl Zeiss 510 Confocal Microscope using a 40x oil objective at RFP/DAPI filter for TMRM and Hoechst respectively, while maintaining the cells at 37°C and in a humidified atmosphere with 5 % CO2. Images were quantified using NIH ImageJ software (https://imagej.nih.gov/ij/). Mean integrated intensities of images (20 randomly chosen fields) in the red channel were determined after background subtraction and the results were presented in graph forms.
2.13. Measuring the reactive oxygen species (ROS) production
ROS generation was measured using mitochondrial superoxide sensitive fluorophore MitoSOX Red kit (Molecular Probes, catalog no. M36008). The SVG-A and PHFA cells were grown in 6-well tissue culture plates (Corning, catalog no. 3506) and were transduced with either Ad-null or Ad-Agno WT (10 PFU/cell). At 18 h post-transduction, the transduced cells were transferred to 35-mm glass bottom poly-L-lysine coated plates (MatTek, catalog no. p35GC-14-C) and incubated for 5 hrs to allow the cell adhesion and were then treated with MitoSOX Red following the manufacturer’s recommendations. In brief, the culture medium was removed, and the transduced cells were treated with DMEM containing MitoSOX Red (5 uM) and counterstained with Hoechst 33342 (5 μg/ml) for 15 min 37°C in a humidified atmosphere with 5 % CO2. After staining, cells were washed twice with 1xPBS and the live cells were imaged using a Carl Zeiss 510 Confocal Microscope using a 40x oil objective at RFP/DAPI filter for MitoSOX Red and Hoechst respectively, while maintaining the cells at 37°C in a humidified atmosphere with 5 % CO2. Images were quantified using NIH ImageJ software (https://imagej.nih.gov/ij/). Mean integrated intensities of images (from 20 randomly chosen fields) in the red channel were determined after background subtraction and results were presented in graph forms.
2.14. Replication assay
Replication assays were carried out as previously described with minor modifications (Sariyer et al., 2011). Briefly, the plasmid constructs (Bluescript KS-JCV Mad-1 WT and Bluescript KS-JCV Mad-1 Agno (15-71) were digested with BamHI to liberate the viral genomic DNA from the vector (Bluescript KS+). SVG-A cells (2x106 cells/75cm2 flask) were then separately transfected/infected with BamH1 digested DNA (10 μg each) using Lipofectamine™ 3000 according to manufacturer’s recommendations. Next day, cells were transferred into 175cm2 tissue culture flasks and fed with 30 ml complete DMEM media supplemented with 10 % FBS. Half of the media were then replenished every three days posttransfection until termination of the infection cycles for each sample. At the indicated time points, low-molecular-weight DNA containing both input and replicated viral DNA was isolated as follows: Cells were harvested by trypsinization, washed with PBS and pelleted by centrifugation (600 g for 3 min). Cells were then resuspended in Qiagen P1 buffer (250 μl, 50 mM Tris-HCl pH 8.0, 10 mM EDTA, 100 μg/ml RNaseA), lysed with Qiagen P2 buffer (250 μl, 200 mM NaOH, 1% SDS) and neutralized with Qiagen P3 buffer (250 μl, 3.0 M potassium acetate pH 5.5). The cell lysates were centrifuged for 4 min at 10,000 g and the supernatants (650 μl) were transferred into clean Eppendorf tubes without taking any cell debris. DNA was precipitated by 100% isopropanol (650 μl) by centrifuging the samples at 10,000 g for 15 min at room temperature. Each DNA pellet was resuspended in 100 μl dH2O containing RNase A (1 μg total/sample). Twenty five microliters of DNA samples were digested with BamHI and DpnI enzymes in 1xCutSmart NEB buffer for 3h at 37 °C, resolved on a 1% agarose gel, and transferred onto nitrocellulose membrane (Amersham hybond-N, catalog no. RPN 203N). Southern blots were analyzed using JCV-specific probes as describe previously (Sariyer et al., 2011).
3. Results and Discussion
3.1. Agnoprotein harbors a mitochondrial targeting sequence (MTS) and localizes to mitochondria
Viruses are obligatory intracellular parasites, most of which encode a limited number of regulatory proteins that are not sufficient to complete their life cycle. Therefore, they target and highjack various cellular host proteins and organelles though their regulatory proteins to create a more conducive environment for the completion of their propagation cycle. Such regulatory viral proteins often contain various functional domains, such as NLS, NES and MTS, to amplify their targeting and functional power; and direct them to many different cellular components and organelles. For example, the Borna disease virus X protein is such a viral protein which localizes to the nucleus and mitochondria of the infected cells where it exerts its functions accordingly (Poenisch et al., 2009). With respect to organelle targeting, there are a number of viral proteins reported to target mitochondria regardless of the origin of their DNA or RNA genome (Anand and Tikoo, 2013).
All mitochondria targeting proteins contain a specific mitochondrion targeting sequence (MTS) located either in the N-terminus or internal regions of the respective proteins. The N-terminal MTS of the protein is cleaved in the matrix following its import into the mitochondria and then distributed to the various regions of the organelle. The N-terminally located MTS has, in general, several interesting features. (i) it constitutes the first 10-90 amino acid of the protein, (ii) adopts an amphipathic α-helix conformation (von Heijne, 1986) and (iii) this α-helix contains positively charged surface residues mostly made up of Args but no negatively charged residue is allowed on this surface (Schneider et al., 1998; von Heijne, 1986). Finally, MTS also contains a cleavage site at which it is cleaved by “mitochondrial processing peptidase (MPP)” (Schneider et al., 1998) in the matrix compartment of mitochondria.
Agnoprotein, an important regulatory protein of JCV (Sariyer et al., 2006; Sariyer et al., 2011), is known to contain a number of functional domains, including NLS and NES (Saribas et al., 2018a). Additionally, bioinformatics predictions revealed that it also contains another important functional domain, mitochondrial targeting sequence, MTS, located at its N-terminus (aa 1-14, MVLRQLSRKASVK) with having a probability scale closer to 1 (0.9277) as determined by MitoProt program (Claros and Vincens, 1996) (https://ihg.gsf.de/ihg/mitoprot.html). Moreover, recent 3D structure of Agno also revealed that it contains a short amphipathic α-helix domain rich in positively charged Arg residues within its MTS domain (Fig. 1A) (Coric et al., 2017). Furthermore, our proteomics studies revealed that Agno targets a number of mitochondrial proteins including Tom and Tim complexes (Saribas et al., 2020). All these findings are consistent with the above-mentioned common structural features of MTS. Thus, these observations compelled us to further investigate the functional consequences of targeting of mitochondria by Agno.
Fig. 1. Agno contains a mitochondrial targeting sequence.
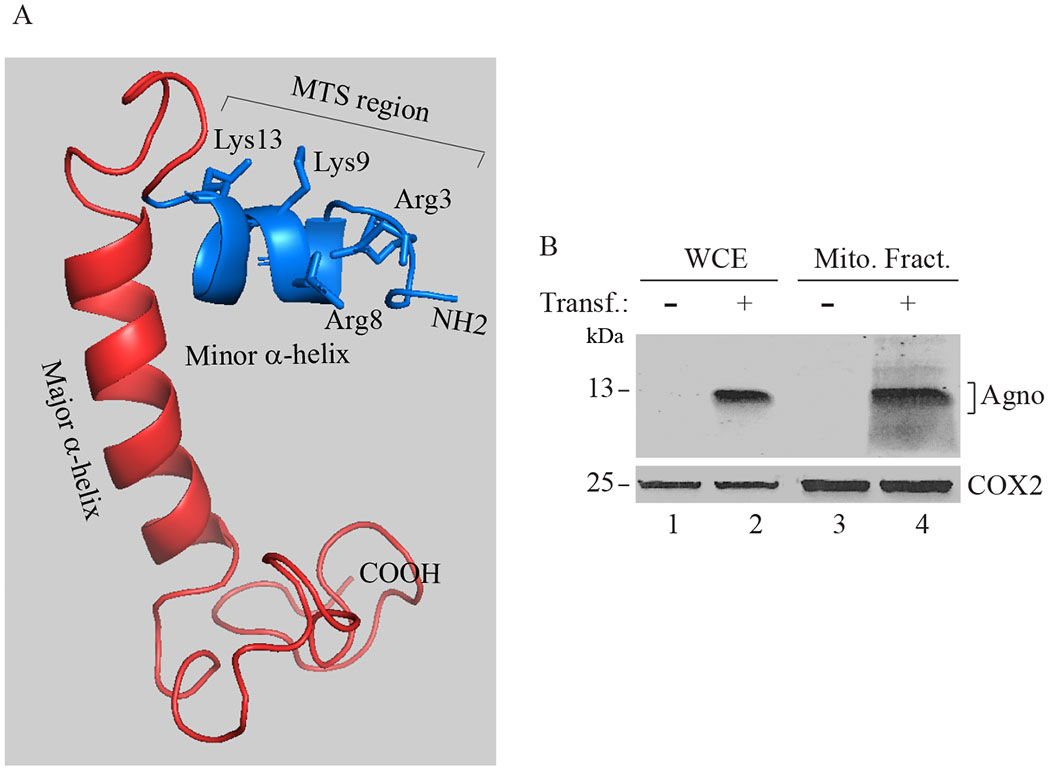
(A) Three dimensional (3D) structure of Agno (Coric et al., 2017). Agno protein contains a minor and a major α-helical region as indicated. The rest of the protein adopts an intrinsically unstructured conformation (Coric et al., 2017; Coric et al., 2014). The N-terminus of Agno is predicted to contain a mitochondria-targeting sequence (MTS) (aa 1-14, MVLRQLSRKASVK). This prediction was made by using MitoProt program (https://ihg.gsf.de/ihg/mitoprot.html), which showed that the probability of mitochondria targeting by Agno is closer to the to 1 (0.9277) (Claros and Vincens, 1996). (B) Mitochondria fractions contain Agno. HEK293T cells were transfected with a pCGT7-Agno expression plasmid and mitochondria was isolated from the untransfected and transfected cells; and analyzed by Western blotting using α-Agno (Del Valle et al., 2002) and α-COX2 (COX2 = Cytochrome C oxidase II, mitochondrial protein) (Abcam, catalog no. Ab3298) antibodies. Whole-cell extracts (WCE) prepared from untransfeced (lane 1) and transfected cells (lane 2) were loaded as negative and positive controls. Transf: Transfection. Mito. Fract.: Mitochondrial fraction.
First, to provide further experimental evidence that Agno actually targets mitochondria, the HEK293T cells were transfected with an Agno-expression plasmid and at 36h posttransfection, mitochondrial fractions were isolated from the transfected and non-transfected cells and analyzed by Western blotting. As shown in Fig. 1B, indeed, Agno was readily detectable in mitochondrial fractions in Agno-positive cells, clearly indicating its co-localization with mitochondria (Fig. 1B, lane 4).
3.2. Agno co-localizes with mitochondrial network
The co-localization of Agno with the mitochondrial network was further examined by immunocytochemistry in Agno-positive SVG-A cells. The SVG-A cell line is an established human astrocytic cell line (Major et al., 1985) which supports the JCV replication cycle. SVG-A cells are in fact morphologically larger than HEK293T cells and thus provide much clearer microscopic information regarding Agno co-localization with mitochondrial network. Also note that Agno is a predominantly cytoplasmic protein, mostly accumulating at the perinuclear area of the cells, although a small portion of the protein is also consistently detected in the nucleus (Saribas et al., 2012). SVG-A cells were then transfected with an Agno-expression plasmid, processed for immunocytochemistry using α-T7 (monoclonal) and α-COX4 (polyclonal) antibodies and examined under a fluorescence microscope. COX4 (cytochrome c oxidase subunit 4) is a mitochondrion specific protein. As shown in Fig. 2, the microscopic images demonstrate a strong co-localization of Agno with the mitochondrial network, as evidenced from the development of a yellow color at the co-localized areas due to the overlap of the red (Agno) and green (COX4) colors.
Fig. 2. Analysis of Agno targeting to mitochondria by immunocytochemistry and Western blotting.
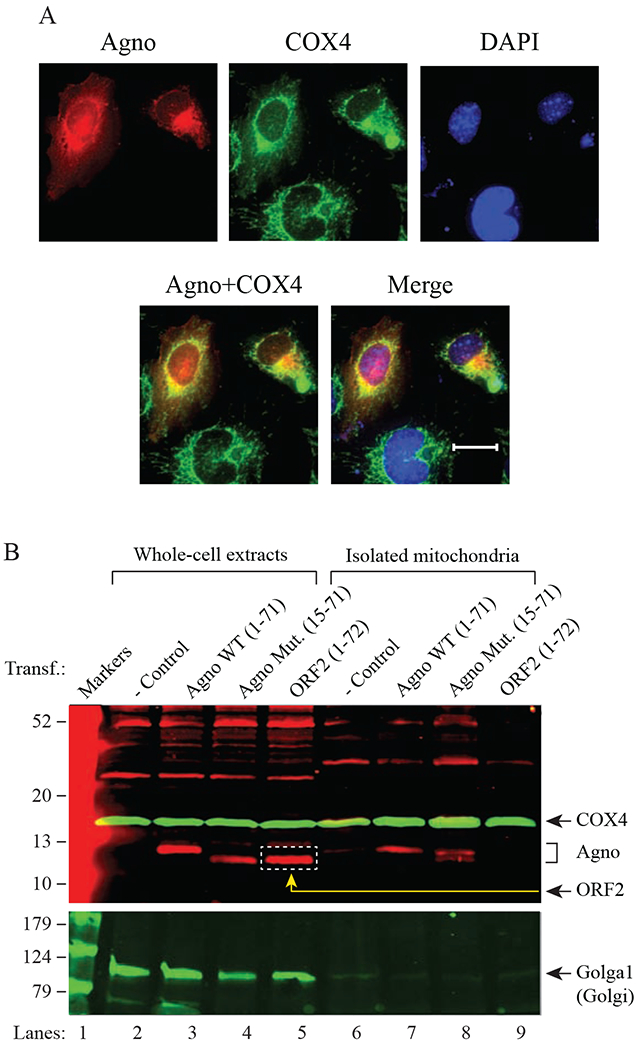
(A) SVG-A cells were transfected with a pCGT7-Agno expression plasmid and processed for immunocytochemistry at the 24h posttransfection using α-T7 (monoclonal, EMD Millipore, catalog no. 69522) and α-COX4 (polyclonal, GeneTex, GTX101499) antibodies as described in materials and methods. Scale bar: 22 μm. (B) Analysis of the purified mitochondrial samples prepared from untransfected and transfected cells by Western blotting. HEK293T cells, untransfected or transfected with either pCGT7-Agno WT, pCGT7-Agno (15-71) mutant or pCGT7-ORF2 (Saribas et al., 2018a) were subjected to either the whole-cell extract (WCE) preparation or to the mitochondria purification using α-TOM22 antibody coupled to magnetic beads as described in materials and methods. Samples were then analyzed SDS-4-20% PAGE followed by Western blotting using α-T7 to detect T7-Agno WT, T7-Agno (15-71) mutant and T7-ORF2 (negative control). Western blots were also probed with α-COX4 (polyclonal, mitochondria marker, GeneTex, catalog no. GTX101499) and α-Golgin-97 (monoclonal, to detect Golgin1, Golgi marker, Cell Signaling, catalog no. 13192). In lanes 2-5, 40 μg of WCE were loaded. In lanes 7-10, 1/5 of the purified mitochondria was loaded on the gel. Transf.: Transfection.
3.3. Mitochondrial targeting by Agno was also analyzed in affinity purified mitochondrial fractions by Western blotting
Western analysis of the mitochondrial fractions isolated by a detergent-based strategy provided evidence that Agno indeed co-fractionates with mitochondria (Fig. 1B). However, such an analysis did not exclude the possibility that Agno detection on Western blots is due to a contamination by the other organelles that Agno also targets in cells such as the Golgi apparatus. In order to eliminate such a possibility, we employed a new purification method, called “antibody-based (affinity-based) purification of mitochondria (Afanasyeva et al., 2018; Franko et al., 2013), which is an elegant strategy and utilizes an α-TOM22 (TOM22 is an outer membrane protein) antibody coupled to magnetic beads. It captures only the mitochondria when applied to a strong magnetic field to the magnetic beads as described in material and methods. To achieve this, HEK293T cells were transfected with either Agno WT or Agno (15-71) mutant (MTS deleted mutant) or ORF2 (a 72 aa long and unrelated short protein with no MTS) (Saribas et al., 2018a) expression plasmids and mitochondria were affinity purified using α-TOM22 antibody (Afanasyeva et al., 2018; Franko et al., 2013) from those transfected cells. The purified mitochondria were analyzed by Western blotting using various antibodies to detect Agno WT, Agno (15-71) mutant, ORF2, COX4 (mitochondrion inner membrane protein) and Golga1 (Golgi marker). As shown in Fig. 2B, Agno WT, Agno (15-71) mutant and ORF2 are readily detectable in transfected cells by α-T7 tag antibody (lanes 3-5). In purified mitochondria fractions, however, we only detected Agno WT and Agno (15-71) mutant (lanes 7-8) but not ORF2 (lane 9), consistent with our findings from the mitochondrion isolation experiments described in Fig. 1B. As predicted, ORF2 with no MTS sequence did not target mitochondria. We also made an interesting observation where Agno (15-71) mutant with no MTS sequence was also be able to co-fractionate with mitochondria (lane 8) suggesting that other sequences within Agno also play a role in its mitochondrial localization in addition to the contributions provided by its MTS sequence. We believe that the prime suspect for such sequences lies within the dimerization domain of Agno (aa 24-39) (Coric et al., 2017; Saribas et al., 2013; Saribas et al., 2011), because such a region was recently assigned to the dimerization domain of the BK virus (BKV) agnoprotein (Manzetti et al., 2020). As we predicted, we did not detect Golgi1 on the purified mitochondria fractions as readily as those for Agno WT or its mutant, although it is clearly detectable on lanes loaded with WCEs prepared from both the untransfected and transfected cells (lanes 2-5), providing clear evidence against a possibility that Agno detected on isolated or affinity-purified mitochondria fractions in fact comes from the contaminated organelles such as Golgi apparatus that Agno also targets (Saribas et al., 2020). As a positive control, COX4 is also detectable in lanes loaded with WCEs (lanes 2-5) as well as on those loaded with purified mitochondrial fractions (lanes 6-9).
We have further examined whether the MTS sequence of Agno would be able to direct any protein to mitochondria when it is fused with, such as GFP. Then, Agno WT (1-71), Agno MTS (1-14) and Agno with no MTS (15-71) were separately fused to GFP protein, transfected into SVG-A cells separately and examined under a fluorescent microscope for their mitochondrial targeting ability as described under materials and methods. Both the GFP-Agno WT and GFP-Agno (15-71) fusion proteins showed a patchy distribution although Agno WT is known to show more even distribution pattern in transfected or infected cells (Saribas et al., 2016). This is because such a fusion most likely triggered a rapid dimerization of Agno, since Agno is known to form stable homodimers and oligomers in vivo and in vitro (Saribas et al., 2013; Saribas et al., 2011; Saribas et al., 2016). Nonetheless, both fusion proteins showed mitochondrial distribution (Supplement 2), which is consistent with our mitochondrial purification and fractionation studies, where we detected the Agno (15-71) mutant in the mitochondrial fractions (Figs. 1B, 2B and 3B). The GFP-Agno (aa 1-14, with MTS) mutant, on the other hand, showed a relatively smoother distribution throughout the cells while also targeting to mitochondria but not to the extent of Agno WT or Agno (15-71) mutant. These observations suggest that Agno MTS contributes to Agno targeting to mitochondria rather than being a strong “bone fide” MTS signal of Agno. In addition, Agno MTS region of Agno was also previously reported to be involved in the viroporin activity of the protein (Suzuki et al., 2010) Taken together, these comprehensive studies establish that Agno targets mitochondria but its detection in purified mitochondrial fractions is not due to a contamination from other organelles such as Golgi that Agno also targets.
Fig. 3. Isolation of mitochondria; and treatment with digitonin and proteinase K.
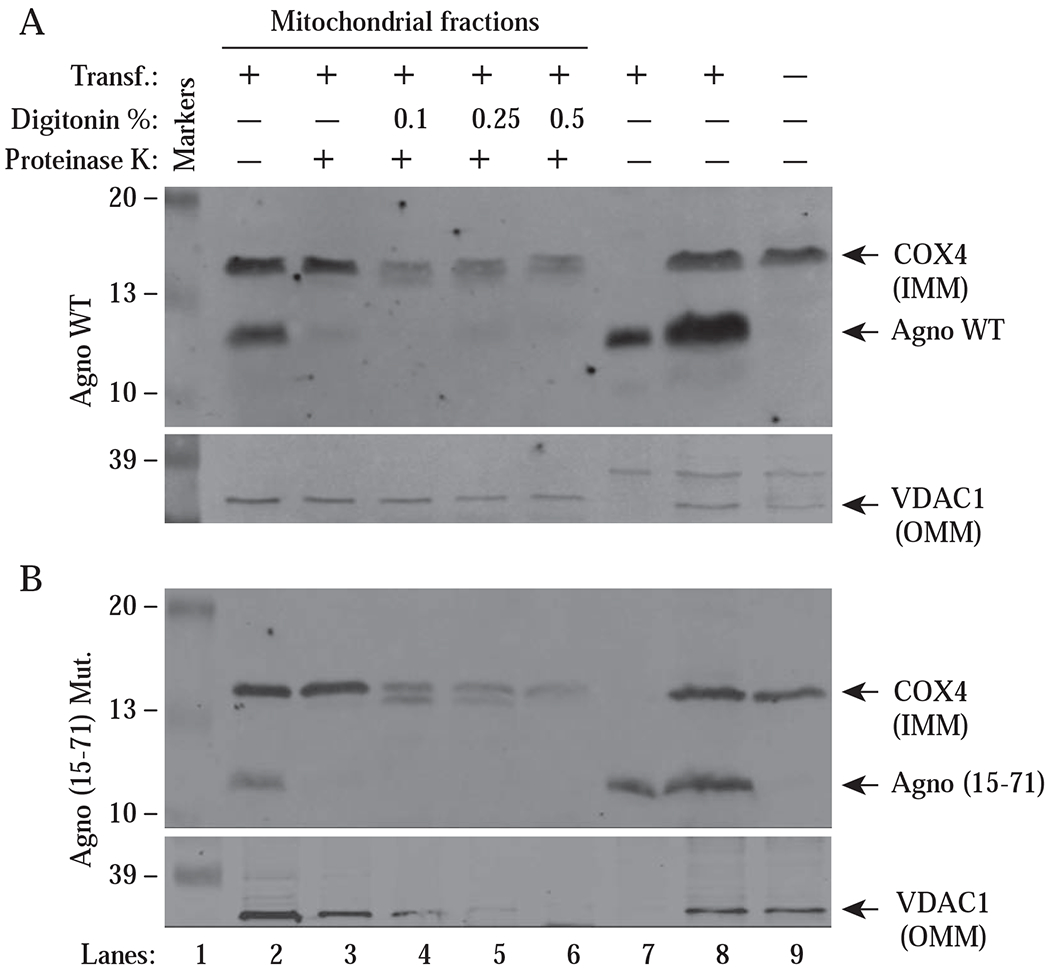
HEK393T cells were transfected with either pCGT7-Agno WT or pCGT7-Agno (15-71) mutant by means of the calcium phosphate precipitation method. At 30 h posttransfection, mitochondrial fractions were prepared from the untransfected and transfected cells, treated with proteinase K in the absence or presence of the increasing concentration of digitonin as indicated on the panel for 15 min and samples were then analyzed by Western blotting using α-Agno, α-VDAC1 and α-COX4 antibodies as described in materials and methods. In lane 7, cytosolic fractions from the transfected cells were loaded onto SDS-PAGE. In lane 8, WCEs from the transfected cells were loaded as a positive control. In lane 9, WCEs from the untransfected cells were loaded as a negative control. COX4 is an integral inner membrane protein of mitochondrion. VDAC1 is an integral outer membrane protein of mitochondrion. Transf.: Transfection, Mut.: Mutant
3.4. Agno co-fractionates with the mitochondrial outer membrane proteins
We have so far provided experimental evidence that Agno targets mitochondria, but these results were not sufficient to dissect out the mitochondrial components to which Agno is localized. To investigate this, we transfected HEK293T cells with either an Agno WT or Agno (15-71) mutant (with no MTS sequence) expression vector. At 30 h posttransfection, mitochondria were isolated, sequentially permeabilized with digitonin (a nonionic detergent that permeabilize mitochondrial membranes) and protein cleavage is performed using proteinase K as indicated on the Fig. 3AB and samples were finally analyzed by Western blotting. The mitochondrial proteins, COX4 (an integral inner membrane protein) and VDAC1 (an integral outer membrane protein) were used as positive controls in the experiments. As shown in Fig. 3, lane 2, Agno was found to co-fractionate with mitochondria, which is consistent with our findings from Figs. 1B and 2B. In addition, the treatment of the isolated mitochondria with proteinase K even in the absence of digitonin resulted in almost a complete degradation of Agno WT protein by proteinase K (~ 90% of Agno WT is degraded) (Fig. 3A, lane 3, upper panel). However, the degradation of Agno (15-71) Mut protein was more dramatic (a complete degradation) (Fig. 3B, lane 3, upper panel). It was surprising to observe that the removal of its MTS sequence did not prevent Agno from localizing to mitochondria. In addition, the densitometric analysis of the bands revealed that Agno (15-71) Mut expresses less efficiently (~21 % less) than Agno WT in cells but its mitochondrial targeting ability is similar to WT (~25 % vs 17 % respectively) suggesting that the remaining regions of Agno may also play a role in its localization to mitochondria. As mentioned above, we believe that the prime suspect for such a region lie within the dimerization domain of Agno (aa 24-39) (Coric et al., 2017; Saribas et al., 2013; Saribas et al., 2011) because a similar potential dimerization domain within BKV agnoprotein was recently identified by Manzetti et al., (Manzetti et al., 2020) as the region contributing to the mitochondrial localization.
3.3. Analysis of the effect of Agno on ATP production in HEK293T cells.
Cells continuously transfer the chemical bond energy stored in sugars, proteins and fats into the most usable and transferable energy form, ATP. The energy transfer takes place in either the cytosol by glycolysis (using no oxygen in the process) or in the mitochondria by oxidative-phosphorylation (utilizing oxygen and producing carbon dioxide and water) processes. Viral proteins targeting mitochondria affect various mitochondrial functions including oxidative phosphorylation (OXPHOS). As such, we next examined the effect of Agno on ATP production in Agno-positive cells by employing a bioluminescence assay, which utilizes the following elements to produce a measurable light: (a) the existing ATP in the samples, (b) a recombinant firefly luciferase and (c) D-luciferin (substrate for luciferase). To assess the effect of Agno on ATP production in cells, HEK293T cells were transfected with either an Agno WT (1-71), Agno (15-71) mutant or ORF2 WT (Saribas et al., 2018a) (a 72 aa long unrelated protein with no MTS, used as a negative control) expression plasmids or vector alone (another control) in triplicates. At 24 h posttransfection, cells were lysed in lysis buffer and samples were then analyzed for ATP production, employing a luciferase assay. Results were then compared graphically as shown in Fig. 4A, which demonstrates that the luciferase activity in samples obtained from the Agno WT and Agno (15–71) mutant transfected cells significantly reduced but not in ORF2 expressing cells implying that both Agno WT and Agno mutant, but not ORF2 significantly altered the ATP production in transfected cells. This shows that both Agno WT and Agno (15-71) mutant have a negative effect on ATP production, which correlates with our findings from Fig 2B and 3B, where we found that both Agno (15-71) mutant and Agno WT target mitochondria. As predicted, although ORF2 has virtually the same amino acid length and exhibits a similar expression level as Agno WT in transfected cells, it did not show a significant effect on the ATP production, emphasizing the fact that expression of an unrelated protein does not necessarily have the same or similar effects on the function of mitochondria as Agno has.
Fig. 4. Indirect analysis of ATP levels in Agno-positive cells by bioluminescence assay.
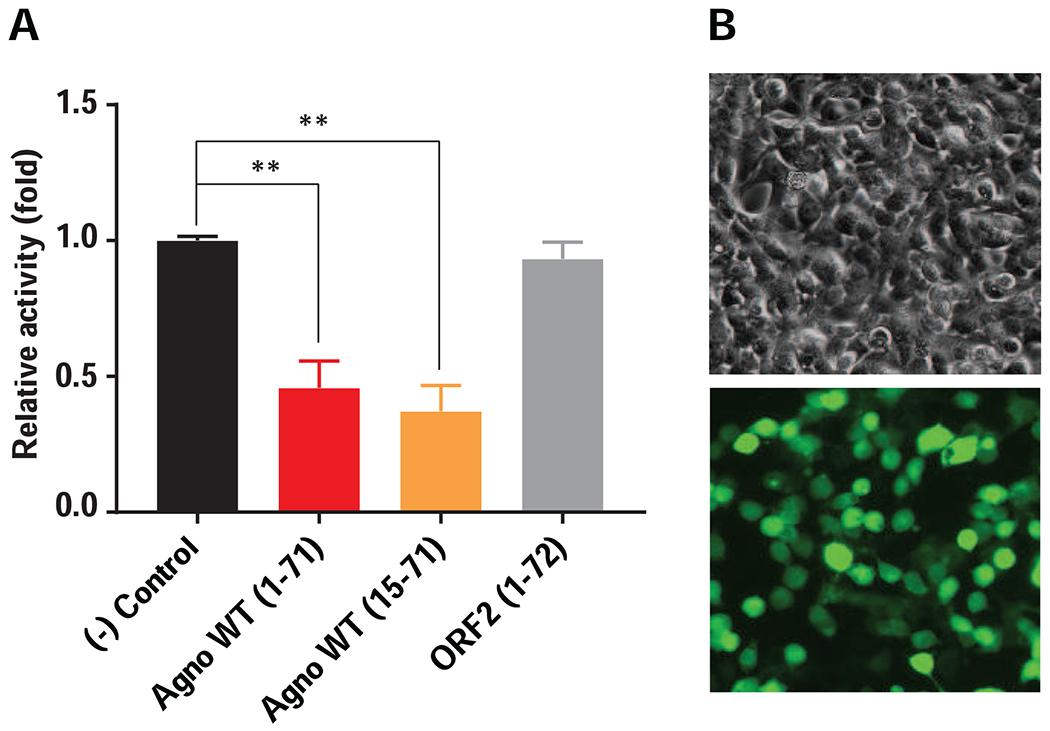
(A) HEK293T cells were transfected with either vector alone [pCGT7, (-) Control], pCGT7-Agno WT, pCGT7-Agno (15-71) mutant or pCGT7-ORF2 expression plasmids in triplicate and at 24h posttransfection, cells were processed for bioluminescence assay to indirectly measure the ATP levels in Agno-positive cells as described in materials and methods. (B) HEK293T cells are highly transfectable by calcium-phosphate method. (Upper panel) Phase-contrast microscopic image of HEK293T cells. (Lower panel) Immunofluorescence image analysis of Agno-transfected cells using α-Agno antibody, which are used in bioluminescence assay.
In addition, we also examined whether the observed negative effect of Agno on ATP production result from a possible apoptotic effect of Agno on the cells rather than its measurable effect on mitochondria. To address this possibility, HEK293T cells were separately transduced by Ad-Agno-WT and Ad-Agno (15-71) mutant viruses in duplicate and live cells were examined for apoptosis at two different time points (at 24h and 48h post-transduction) by two different assays - phase microscope and flow cytometry - as detailed under Supplement 3. Results showed no apparent sign of apoptotic effect of Agno WT and Agno (15-71) mutant on HEK293T cells under our short-term assay conditions. However, the apoptotic effect of Agno on long term cultures was previously reported by Merabova et al., (Merabova et al., 2008). Collectively, these results demonstrate that the observed effect of Agno on ATP production is not due to overexpression of the protein or its apoptotic effect on the cells but rather its functional effect on mitochondria.
3.4. Agno significantly induces Ca2+ influx into mitochondria
Calcium (Ca2+) acts as a second messenger to regulate many cellular processes not only inside the cytoplasm but also in selective organelles, including endoplasmic reticulum (ER) and mitochondria (Berridge et al., 1998). Calcium stored in the ER is imported into the mitochondria through the mitochondria-associated ER membranes (MAMs) or cytoplasmic Ca2+ enters directly to mitochondria. In both cases, however, the Ca2+ import into mitochondria is regulated by mitochondria outer or inner membrane-associated protein channels, which have specific affinity for Ca2+. For example, under ER-stimulated conditions, Ca2+ is released from the ER stores travels through a tetrameric Ca2+ ion channel, which is composed of “Ryanodine receptors” (RyRs) and “inositol 1,4,5-triphosphate (IP3) receptors (IP3Rs), which are located at the junction of the ER-mitochondria contacts (MAMs). Ca2+ is then transferred into mitochondrial intermembrane space through a well-characterized mitochondrial outer membrane (MOM) associated protein called, the voltage-dependent anion channel (VDAC) (Marchi et al., 2017; Van Petegem, 2012). The physical interaction between the “RyRs and IP3Rs” complex and VDAC is mediated by a glucose-regulated protein 75 (GRP75) which plays an important role in Ca2+ transfer through the MAMs to mitochondria (Szabadkai et al., 2006). The accumulated Ca2+ in the mitochondrial intermembrane space then passes through another highly selective Ca2+ channel to reach the mitochondrial matrix. This channel complex is located at the inner mitochondrial membrane (IMM) and is composed of several known proteins, including mitochondrial Ca2+ uniporter (MCU) (De Stefani et al., 2011; Patron et al., 2013), mitochondrial Ca2+ uniporter isoform b (MCUb) (Lambert et al., 2019; Raffaello et al., 2013), (which functions as a dominant negative regulator of MCU), an essential regulator of MCU (EMRE) (Sancak et al., 2013), mitochondrial calcium uptake 1 and 2 proteins (MICU1 and MICU2), (which are the essential gatekeepers of MCU-mediated mitochondrial Ca2+) (Csordas et al., 2013; Marchi et al., 2017; Perocchi et al., 2010; Plovanich et al., 2013), mitochondrial calcium uniporter regulator 1 (MCUR1) (Tomar et al., 2016). All these proteins constitute the mitochondrial uniporter complex (mtCU). Finally, the imported mitochondrial Ca2+ stimulates the allosteric activation of the Krebs cycle enzymes leading to the stimulation of ATP synthesis. However, during the stress conditions, including the viral infection cases, mtCU complex loses the control of its elegant Ca2+ import mechanisms and causes a Ca2+ overload in mitochondria, which then becomes an unmanageable mitochondrial event that eventually leads to activation of the mitochondrial permeability transition pore (mPTP) complex for Ca2+ efflux, loss of mitochondrial membrane potential and cell death (De Stefani et al., 2011).
Several viruses were reported to alter the Ca2+ uptake into the mitochondria for successful completion of their life cycle. For example, Herpes simplex virus-1 (HSV-1) causes a gradual decrease in Ca2+ uptake by mitochondria during its lytic cycle, although mitochondrial Ca2+ uptake fluctuates during the course of a measles virus infection cycle (Lund and Ziola, 1985). The core protein of hepatitis C virus (HCV) targets mitochondria and increases Ca2+ uptake by enhancing the activity of mtCU complex, causing cellular stress and cell death (Baum et al., 2011; Li et al., 2007). The p7 protein of HCV on the other hand forms porin-like structures and increases the Ca2+ influx into cytoplasm from the storage organelles (Griffin et al., 2004). Additionally, towards the end of the poliovirus infection cycle, virus induces the release of Ca2+ from the lumen of ER through the IP3R and RyRs channels and causes the accumulation of Ca2+ in the mitochondria, rendering mitochondrial dysfunction and cellular apoptosis (Brisac et al., 2010). Likewise, it was also shown that pUL37 protein of cytomegalovirus (CMV) targets mitochondria, causes an increase in mitochondrial Ca2+ levels and thereby increases the ATP and virus production (Sharon-Friling et al., 2006).
Our findings, so far, showed that Agno targets mitochondria and negatively affects the energy production. We next sought to investigate whether Agno has any effect on mitochondrial Ca2+ uptake, because the Ca2+ signaling plays critical roles in the function of mitochondria either in stimulating electron transport complexes and therefore ATP production or in the activation of the mPTP complex and thereby loss of mitochondrial membrane potential and finally cell death (Granatiero et al., 2017; Marchi et al., 2017). To investigate the role of Agno on mitochondrial Ca2+ uptake, SVG-A cells were transfected with either control plasmid, pCGT7 or with pCGT7-Agno and at 24 h posttransfection, cells were processed for cytosolic and mitochondrial Ca2+ uptake as described in materials and methods (Doonan et al., 2015). Fluo-4 and Rhod-2 dyes are known to specifically detect cytosolic and mitochondrial Ca2+ respectively. Ionomycin (control) is an ionophore, which facilitates the Ca2+ influx across the plasma membrane regardless of the presence of Agno. The fluorescence intensities of individual Agno-positive cells were measured for Fluo-4- and Rhod-2 in presence or absence of ionomycin. As shown in Fig 5A and 5B, mitochondria Ca2+ levels (both basal and stimulated) significantly increased in Agno-positive cells, revealed by Rhod-2 dye, which clearly demonstrates the induction of Ca2+ influx into mitochondria by Agno. Also, note that the cytosolic and mitochondrial Ca2+ levels were detected to be higher in Agno-positive cells than controls even before ionomycin treatment, further revealing the Agno related alterations in the Ca2+ signaling pathways.
Fig. 5. Agno significantly induces mitochondrial Ca2+ influx.
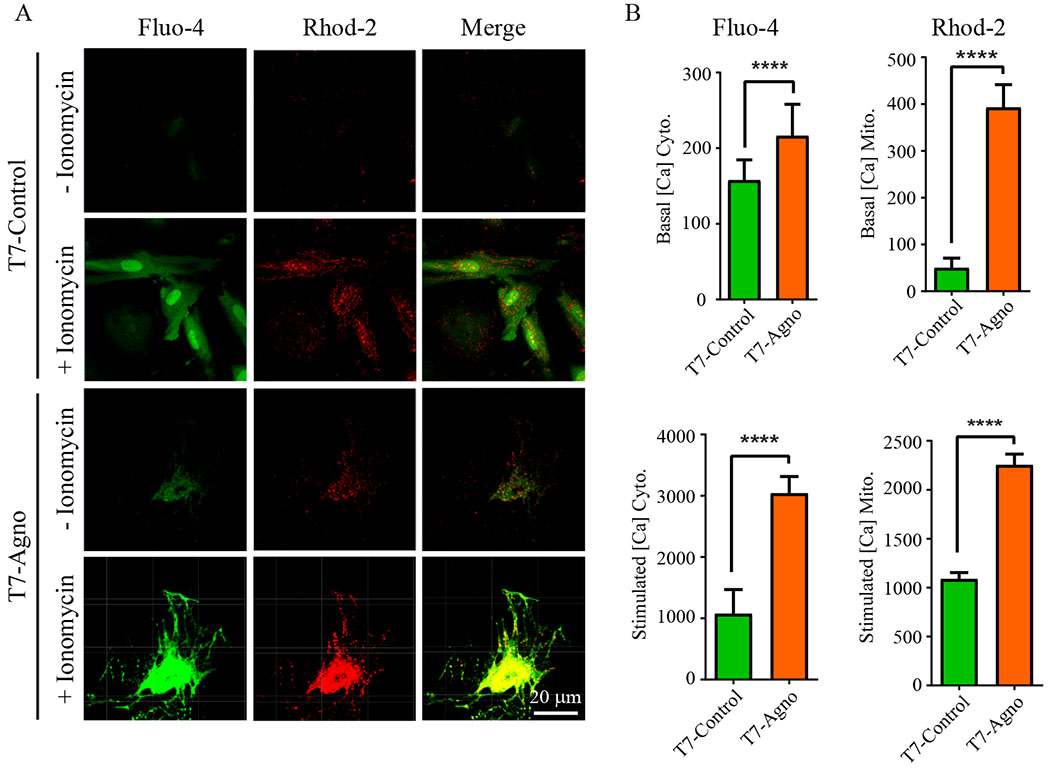
Cytosolic and mitochondrial Ca2+ levels were measured simultaneously in the presence and absence of Agno as described (Doonan et al., 2014, 2015). Briefly, the PHFA cells were transfected with either pCGT7 alone (control) or pCGT7-Agno WT and, at 24h posttransfection, cells were loaded with Fluo-4 (5 μM) and Rhod-2 (2 μM) dyes for 50 min at 37°C. Time-lapse images were recorded (3-s interval) using a Zeiss 510 META confocal microscope with the same illumination and gain settings for all experiments. Live cells were also treated with ionomycin (1 μM) for 15 min to assess the Ca2+ influx into cytosol and mitochondria. (B) Fluorescence intensities were quantified using ZEN 2010 software and presented as the cytoplasmic and mitochondrial Ca2+ detection in graph forms.
3.5. Oxygen consumption rate (OCR) and OCR-related parameters are reduced in Agno-positive cells
Next, we assayed the oxygen consumption rates (OCR) of the Agno-positive cells utilizing a “Seahorse XF26 Extracellular Flux Analyzer”, which continuously measures OCR and proton flux into the cell supernatant overtime. The goal of these measurements was to determine the effect of Agno on several mitochondrial respiration parameters in the presence of sequentially injected various electron transport complex inhibitors as indicated in Fig. 6A and 7A. For example, oligomycin (ATPase, ETC complex V inhibitor) was used to determine the ATP-coupled respiration rates of the cells in the presence or absence of Agno. That is, this inhibitor provides information about the levels of O2 consumption, directly coupled to ATP production (Supplement 1) (Rose et al., 2014). A decoupler, FCCP (carbonyl cyanide p-trifluoromethoxy phenyl hydrazine) dissipates the proton gradient built in the mitochondrial intermembrane space by transporting protons across the inner membrane to the matrix. This process takes place independently from the ATP synthase activity, where the OCR is uncoupled from the ATP production. However, O2 is still utilized by the electron transport chain (ETC) at the maximum rate in order to re-establish the proton gradient at the inner membrane space; thereby it allowed us to measure the maximal respiratory capacity of the cells. Finally, antimycin A, a complex III (cytochrome bc1 oxidoreductase complex) inhibitor, totally inhibits ETC which allows us to measure the non-mitochondrial respiration (Supplement 1). A graphical representation of mitochondrial respiration parameters is shown in Supplement 1.
Fig. 6. Measurement of the oxygen consumption rates (OCR) and the OCR-related parameters in Agno-positive cells.
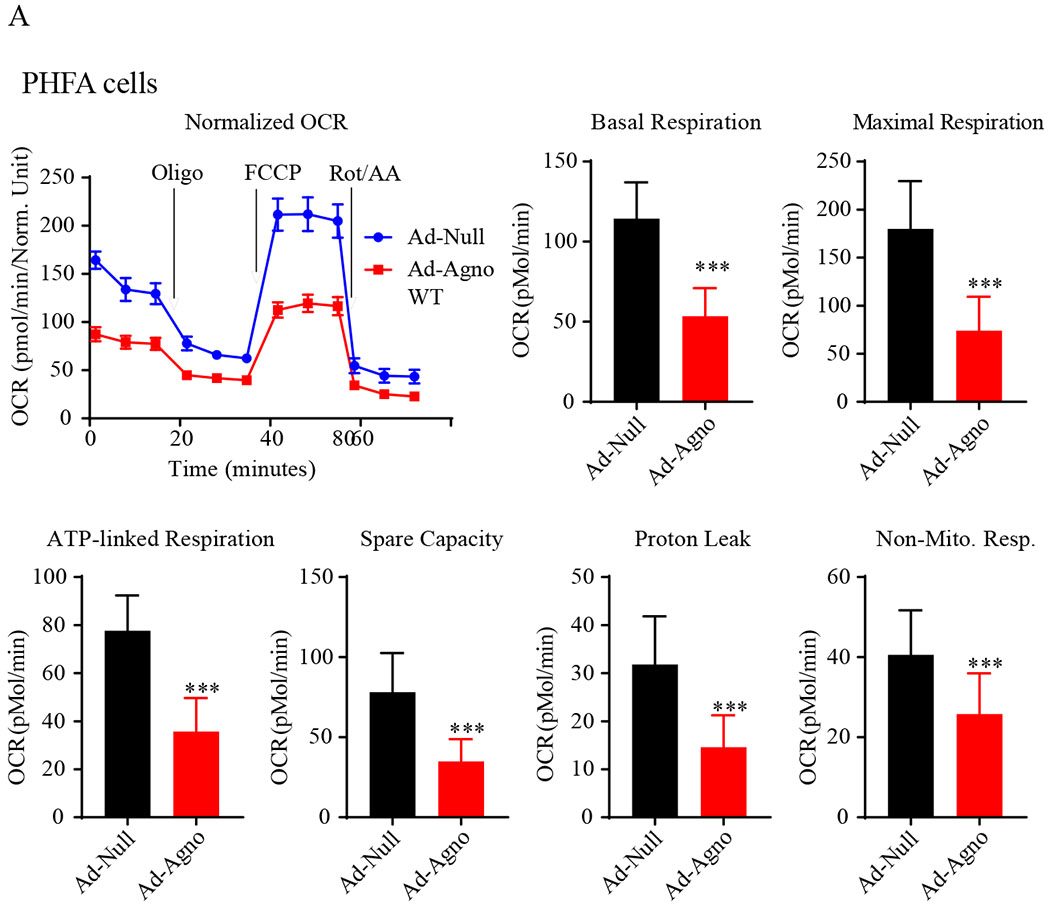
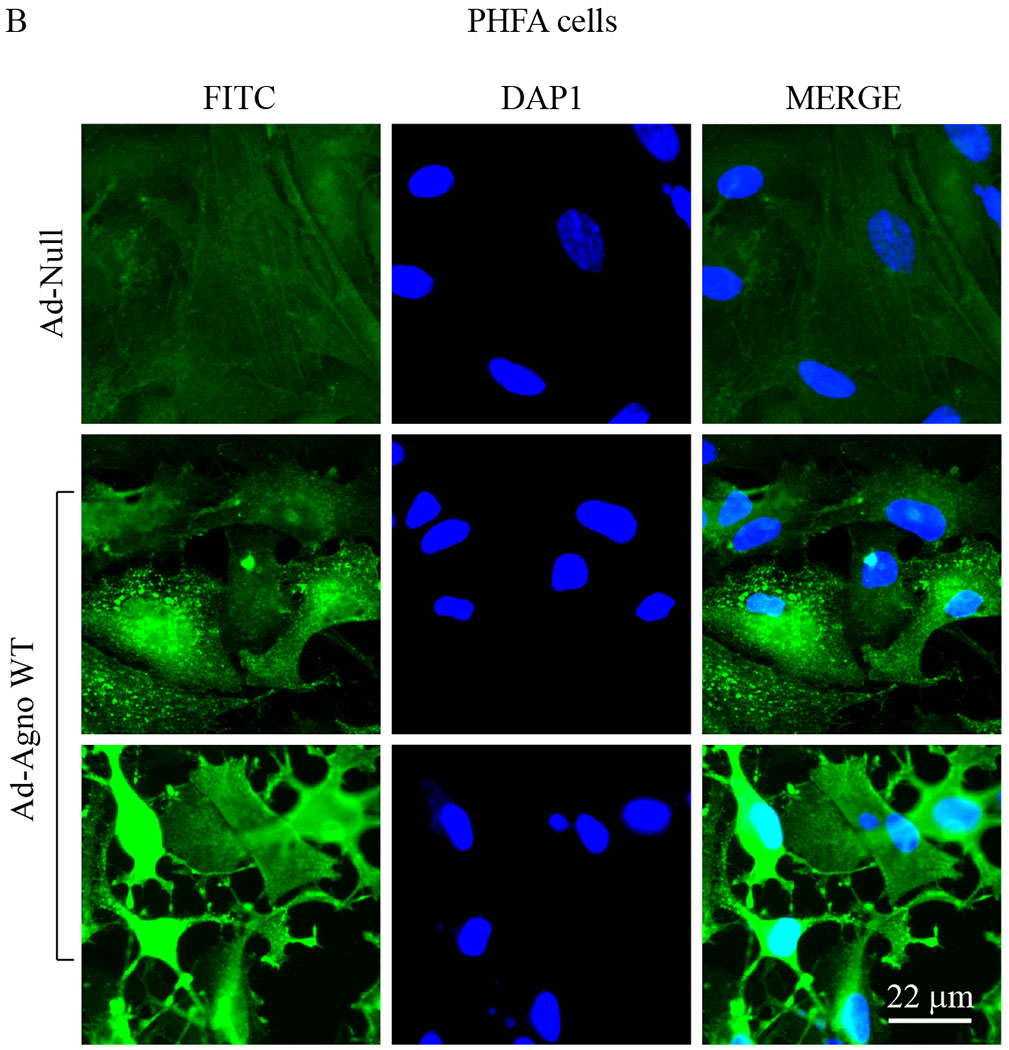
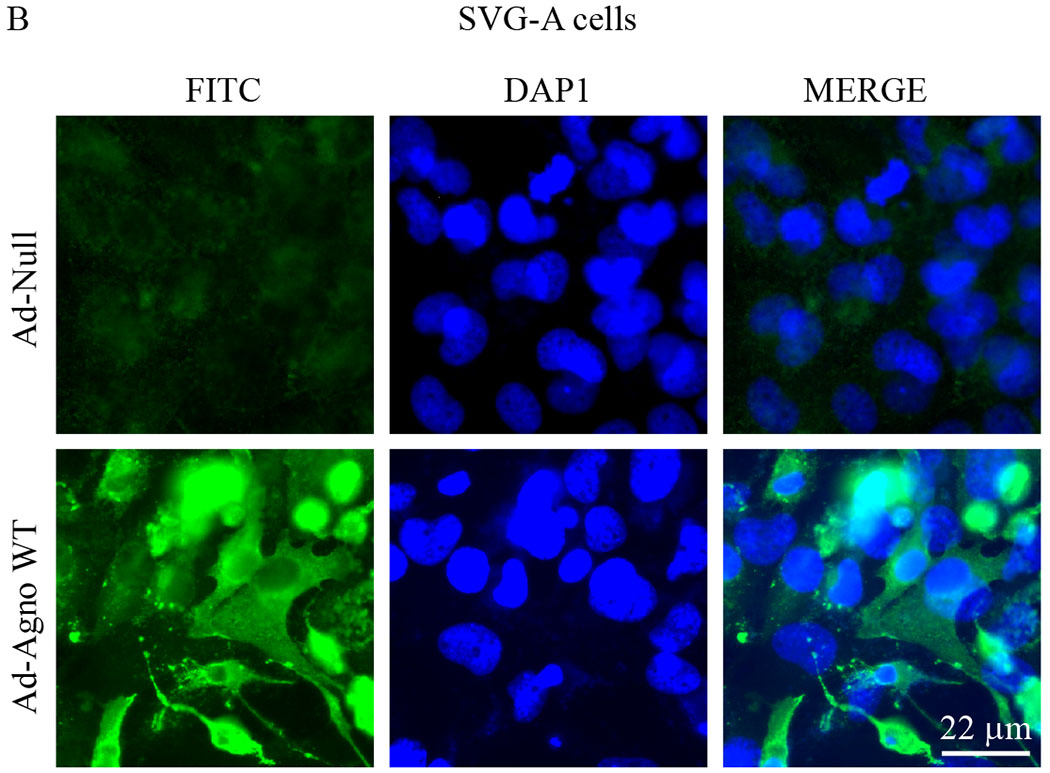
(A) The primary human fetal astrocytes (PHFA) were transduced with either Ad-Null or Ad-Agno WT and oxygen consumption rates (OCR) and OCR-related parameters were determined in the presence of a sequentially injected various mitochondrial inbitors (oligomycin, FCCP, antimycin A/rotenone) using a “Seahorse XF26 Extracellular Flux Analyzer” as described in materials and methods. (B) Ad-Agno-transduced and Ad-Null-transduced PHFA cells were analyzed by immunocytochemistry by probing the cells with a primary polyclonal α-Agno and a secondary fluorescein isothiocyanate (FITC)-conjugated goat α-rabbit antibodies using a fluorescence microscope as described under materials and methods. Scale: 22 μm. Non-Mito. Resp.: Non-Mitochondrial Respiration.
Fig. 7. OCR analysis of Agno-positive SVG-A cells.
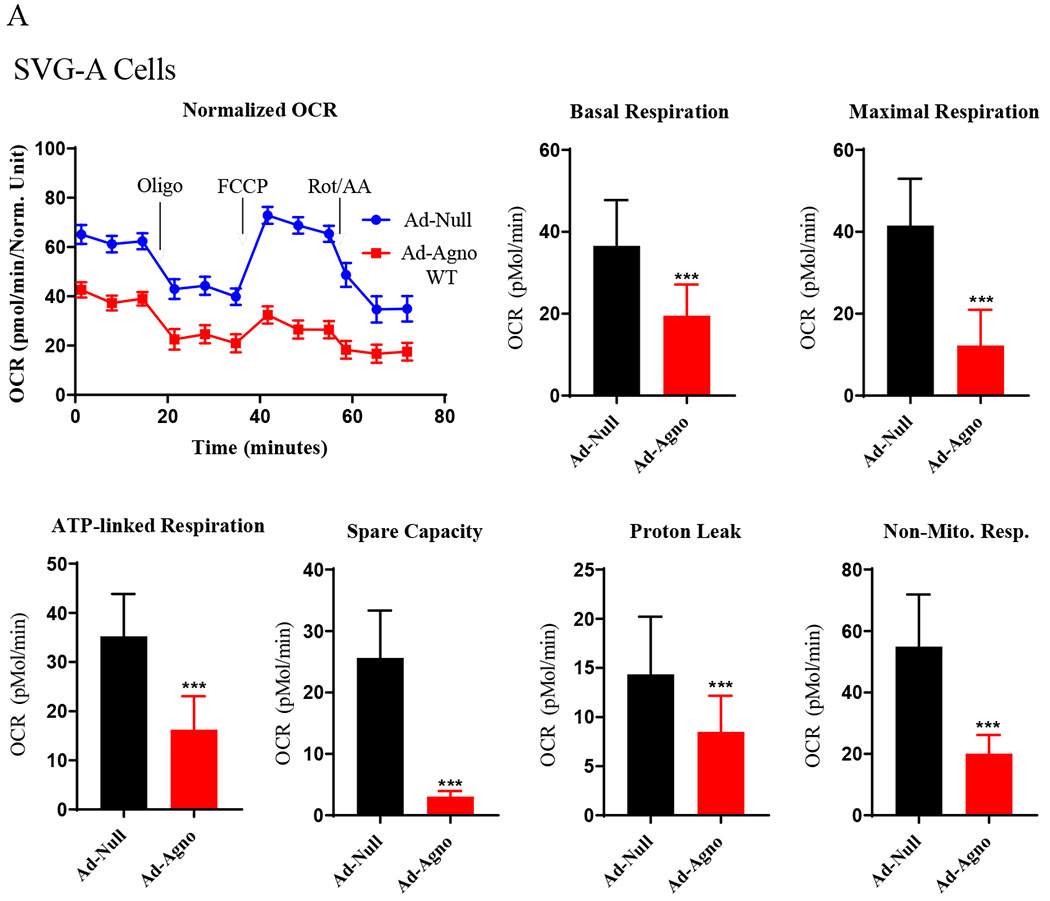
(A) SVG-A cells were transduced with either Ad-Null or Ad-Agno WT viruses and OCR parameters were determined as described under Fig. 6A. (B) In parallel to OCR studies described in panel A, SVG-A cells were also analyzed by immunocytochemistry as described for Fig. 6B using α-Agno antibody. Scale: 22 μm. Non-Mito. Resp.: Non-Mitochondrial Respiration.
In order to measure the mitochondrial respiration parameters, including basal respiration, maximal respiration, ATP-coupled (linked) respiration, spare (reserve) capacity, proton leak and non-mitochondrial respiration, both SVG-A and PHFA cells were transduced with either Ad-Agno or Ad-null viruses as described in materials and methods. At 36 h post-transduction, cells were sequentially treated with various ETC inhibitors as indicated on Fig 6A and Fig. 7A, OCR measurements were made using the “Seahorse XF26 Extracellular Flux Analyzer”. Results clearly showed that Agno significantly decreased the OCR of the both cells lines (Fig. 6A and Fig. 7A). This reduction is reflected not only in the basal respiration levels but also in other OCR parameters, including maximal respiration, ATP-coupled respiration, spare capacity, proton leak and non-mitochondrial respiration, suggesting that Agno affects not only mitochondrial functions but also globally alters other mitochondrial processes in cells. Such a global effect is supported by our recent findings where we have recently mapped the cellular targets of Agno to construct its first interactome. This proteomics study showed that Agno targets various cellular networks including those involved in protein synthesis, degradation and cellular transport etc. and several organelles including mitochondria, nucleus and ER-Golgi network (Saribas et al., 2020). With respect to targeting mitochondria, our proteomics data further revealed that Agno interacts with nearly 50 mitochondrial proteins including those involved in ETC, mitochondrial import pathways, and metabolic enzymes, which corroborates with our current findings regarding a negative regulation of OCR parameters by Agno in cells. With respect to the interaction of Agno with the mitochondrial import pathways, proteomic data also showed that Agno targets TOM70 and TOM22 in the TOM complex and several components of TIM23 complex (Saribas et al., 2020) suggesting that it may dysregulate the mitochondrial functions by targeting its protein import pathways, thereby effecting the OCR of mitochondria. Also note that the efficiency of the transduction of both SVG-A (Fig. 6B) and PHFA (Fig. 7B) cells were analyzed by the immunocytochemistry. In both cases, it was apparent that both cell types were efficiently transduced with Ad-Agno virus.
3.6. The level of “reactive oxygen species” (ROS) is substantially increased in Agno-positive cells
Superoxides are produced due to the premature electron leakage that occurs during the passage of the electrons from one electron transport chain complex to another. When these prematurely leaked electrons react with O2, they generate highly reactive oxygen species which are grouped under two superoxide category, (1) O2 − (in its anionic form) and (2) H2O2 − (in its protonated form). The steady state levels of these reactive oxygen species are regulated by various cellular defense mechanisms. However, under cellular stress conditions, including a viral infection case, cells cannot regulate the unmanageable amount of such species, which would then have a harmful effect on the health of the cells. Such circumstances lead to cell death. Alternatively, these species sometimes act as signals for the activation of various cellular pathways including NF-κB signaling pathway, which in turn help a virus to utilize this chaotic situation to transcribe its genes well and replicate its genome more efficiently (Lassoued et al., 2010; Pyo et al., 2008). Sometimes specific viral proteins can sensitize cells to the reactive O2 species-mediated cell death. For example, Hepatitis B virus X protein was found to induce the pro-apoptotic pathways upon exposure of the cells to the oxidative stress by accelerating the loss of Mcl-1 protein (Hu et al., 2011).
“MitoSOX Red”, fluorogenic dye, is known to selectively targets mitochondria and is oxidized by superoxide produced in this organelle and exhibits a red fluorescence, which can be quantified experimentally. Note that MitoSOX is not oxidized by other ROS or reactive nitrogen species indicating its specificity for superoxide. MitoSOX Red was then used to evaluate the impact of Agno on the ROS production by mitochondria in Agno-positive cells. For this purpose, Agno was expressed in both SVG-A and PHFA cells through either Ad-Agno WT (Fig. 8A) or Ad-Null (Fig. 8C) virus transduction and the cells were then treated with MitoSOX Red dye, and the differential fluorescence intensities between the control and experimental settings were quantified as described under materials and methods. As shown in Fig. 8B and D, compared to controls (non-infected and Ad-Null), Agno WT expression in both cell types (SVG-A and PHFA cells) led to significantly higher levels of ROS production (Fig. 8B and 8D).
Fig. 8. Analysis of the effect of Agno on the production of the reactive oxygen species.
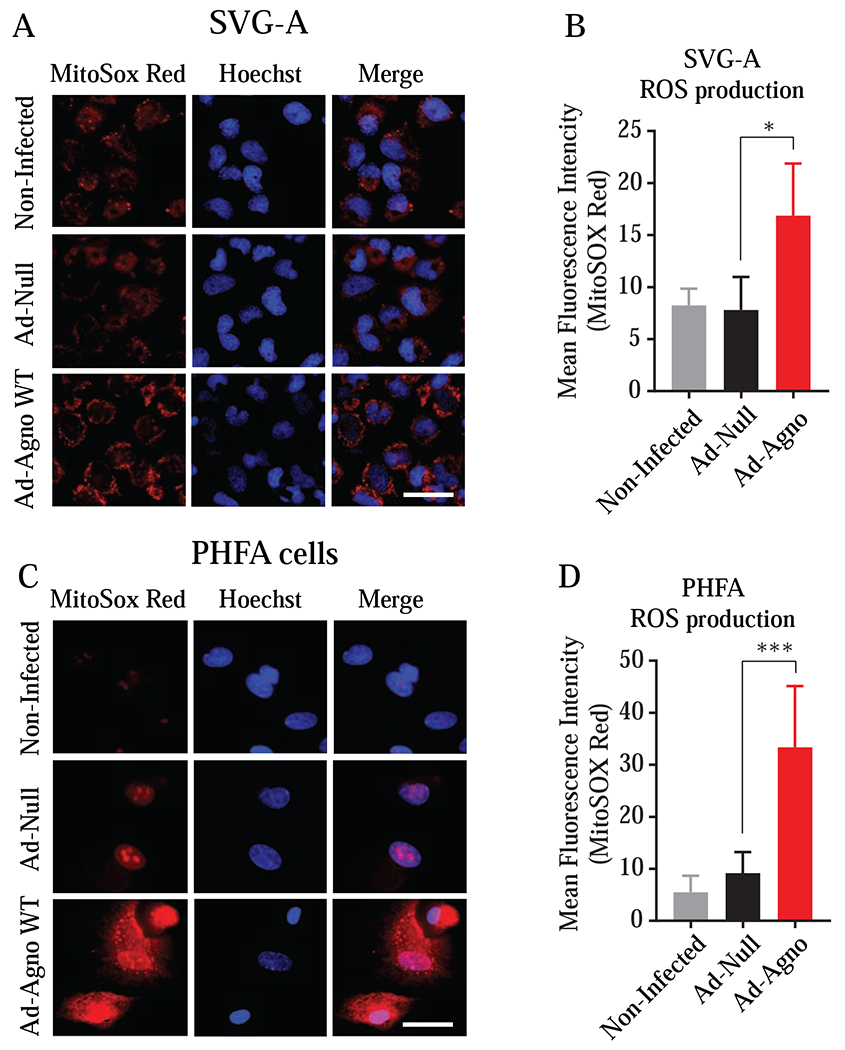
The SVGA (A, B) and PHFA (C, D) cells were transduced with either Ad-Null or Ad-Agno WT viruses. At 18 h post-transduction, the live cells were treated with MitoSOX Red following the manufacturer’s protocols as described in materials and methods. After staining, the cells were imaged using a Carl Zeiss 510 confocal microscope and the fluorescence intensities were quantified using NIH ImageJ software (https://imagej.nih.gov/ij/). Mean integrated intensities of images (from 20 randomly chosen fields) in the red channel were determined after background subtraction and data was presented in graph forms.
3.7. Agno significantly alters the mitochondrial membrane potential (MMP)
Mitochondrial membrane potential is generated because of protons pumped from the matrix to the intermembrane space in an energy dependent manner through the electron transfer complexes located at the inner mitochondrial membrane. These complexes include complex I, III and IV (Zorova et al., 2018). MMP is essential for the maintenance of the electron transport system, ATP production and viability of mitochondria. Excessive oxidative stress, calcium overload and ATP depletion can result in the dissipation of MMP through the formation of mitochondrial permeability transition pores (MPTP) in the inner mitochondrial membrane (Bernardi, 1999). Various viral proteins were reported to manipulate the MMP and mitochondrial ion permeability to successfully complete their life cycle. Hepatitis C virus (HCV) p7 protein, for example, targets mitochondria and regulates the membrane permeability to cations (Griffin et al., 2004) to promote cell survival and thereby viral replication (Gonzalez and Carrasco, 2003). HIV-1 protein Vpr induces the loss of MMP and thereby causes the release of cytochrome c from mitochondria and eventually leads to cell death (Azuma et al., 2006; Jacotot et al., 2000; Zorova et al., 2018). Another HIV-1 protein Tat also induces a reduction in MMP through the modulation of MPTP and eventually leads to a caspase-dependent apoptosis (Macho et al., 1999). Similarly, Influenza A virus PB1-F2 protein targets mitochondria, interacts with voltage-dependent anion channel 1 (VDAC1) and adenine nucleotide translocase 3 (ANT3), reduces the MMP, causes the release of pro-apoptotic proteins and cell death (Chen et al., 2001; Gibbs et al., 2003).
We also examined the effect of Agno on MMP by an indirect measurement assay. The tetramethylrhodamine ethyl ester (TMRE) is a cell permeant and positively charged red orange dye that can stain negatively charged healthy mitochondria and can be used to monitor the MMP levels. The depolarized and inactive mitochondria cannot hold TMRE in live cells. Then to determine the effect by Agno, SVG-A cells were co-transfected with a combination of a GFP plus Agno WT or GFP plus Agno (15-71) mutant expression plasmids and the transfected cells were then subjected to a selection process by flow cytometry based on the expression of GFP in transfected cells. These selected cells were then treated with TMRE and imaged under a florescence microscope (Fig. 9A) as described under the materials and methods section. Fluorescence intensities were quantified, and the results were presentenced in graph form (Fig. 9B). Results showed that Agno significantly reduces the MMP levels (Fig. 9B). Additionally, Agno with no MTS mutant [Agno (15-71)] also showed a considerable negative effect on the MMP, which is consistent with our findings from the mitochondria purification and fractionation assays, where it was observed that Agno (15-71) mutant was also able to target mitochondria (Fig. 2B and 3B).
Fig. 9. Analysis of the effect of Agno on mitochondrial membrane potential.
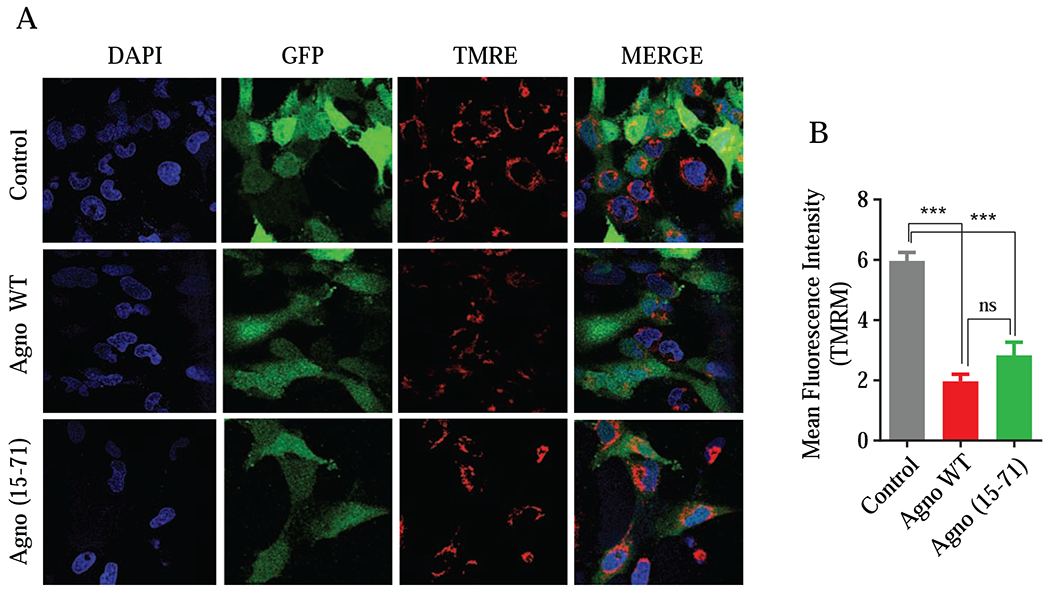
(A) SVGA cells were co-transfected with expression plasmids in the following combination: pGFP-N1 plus pCGT7-Agno WT or pGFP-N1 plus pCGT7-Agno (15-71) mutant in 1:6 ratio using lipofectamine 3000 reagent. At 18 h post-transfection, cells were subjected to flow cytometry to sort GFP-positive cells and subsequently treated with TMRM following the manufacturer’s recommendations as described under materials and methods. The live cells were then imaged by using a Carl Zeiss 510 confocal microscope while maintaining them at 37°C and in a humidified atmosphere with 5 % CO2. (B) Images were quantified using NIH ImageJ software and mean integrated intensities of images (20 randomly chosen fields) in the red channel were determined after background subtraction. Finally, data was presented in a graph form.
3.8. Effect of the mitochondrial targeting sequence deficient mutant of Agno on JCV replication
We next investigated the effect of the Agno mitochondrial targeting sequence on JCV replication. To achieve this, the mitochondrial targeting sequence of Agno was deleted from the viral background and functional consequences of this deletion were then assessed by replication assays. Note that the infection cycle of JCV takes relatively longer time compared to those of BKV and SV40. While BKV and SV40 require 2-3 days to complete their first round of the infection cycle, it usually takes 6-7 days for JCV in a cell culture system. Additionally, it is also our experience working with JCV infection cycle that the scale of the effect of a mutation on the viral replication cycle can be assessed better when, at least, the first two rounds of the viral infection cycle is examined. We then separately transfected/infected SVG-A cells with either JCV WT or its mutant [JCV Agno (15-71)] and the replicated DNA from each experimental condition was isolated at the indicated time points and analyzed by Southern blotting. As shown in Fig. 10A, there is no significant difference between the WT and mutant virus containing Agno (15-71), both of which replicated at similar rates by 5th day posttransfection/infection (lanes 3 and 4). However, the replication level of mutant virus significantly decreased (~ >3 fold) by 15th day posttransfection/infection compared to that of the WT virus (Fig. 10A, compare lane 5 to 6 and Fig. 10B).
Figure 10. Analysis of the replication efficiency of JCV Agno (15-71) mutant virus (with no MTS).
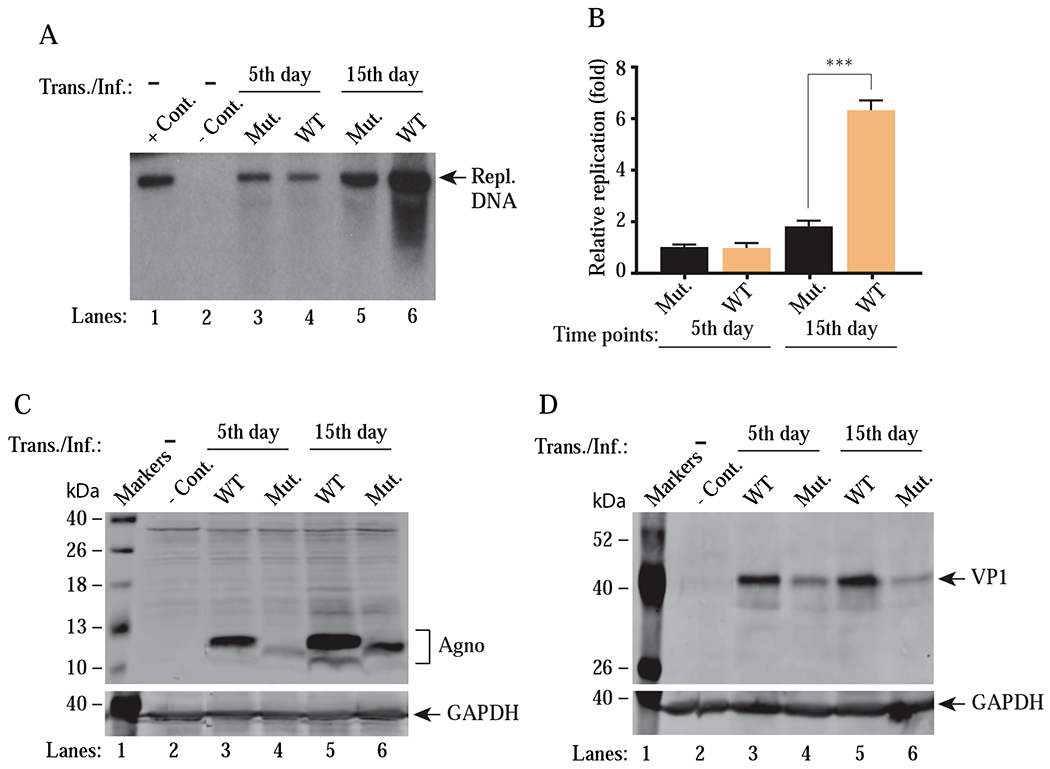
(A) Southern blot analysis of replicated viral DNA. The plasmid constructs [Bluescript KS-JCV Mad-1 WT (WT) and Bluescript KS-JCV Mad-1 Agno (15–71) mutant (Mut.)] were digested with BamHI to liberate the viral genome from the vector and then were separately transfected/infected into SVG-A cells using lipofectamine™ 3000 according to manufacturer’s recommendations. At the indicated time points, the low-molecular-weight DNA containing both input and replicated viral DNA was isolated and digested with BamHI and DpnI restriction enzymes and Southern blot analysis was performed as described in materials and methods. In lane 1, 6 ng of purified JCV Mad-1 WT DNA linearized by BamHI digestion was loaded as positive control (+ Cont.). In lane 2, DNA isolated from the uninfected cells was treated as a negative control (− Cont.). Replication assays were performed in duplicates and a representative data is shown here. (B) Quantitation analysis of Southern blots by a semi-quantitative densitometry method (using NIH Image J program) and presentation of the results in arbitrary units (relative fold replication). Results were statistically analyzed by GraphPad program using One-way ANOVA and data columns were compared by Tukey’s multiple comparison test. *** indicates a significant difference with respect to replication rates between WT and mutant at 15th day posttransfection/infection, (P < 0.0001). (C and D) Western blot analysis of whole-cell extracts prepared from the SVG-A cells, transfected/infected with either WT or mutant [Agno (15-71)] virus to assess the Agno (C) and VP1 (D) levels using α-Agno and α-VP1 (Dugan et al., 2007) antibodies. GAPDH was used as a loading control and detected α-GAPDH antibody (Santa Cruz, catalog no. sc-47724) by Trans./Inf.: Transfection/Infection.
In parallel to replication assays, we also explored the viral protein expression by Western blotting in JCV WT and Agno (15-71) containing mutant virus-infected cells. As shown in Fig. 10C and Fig. 10D, the expression levels of both Agno and VP1 for the mutant virus were noticeably lower than those of WT during the first round of replication (by 5th day) and the these levels did not recover during the second round of replication (by 15th day). Particularly, the relatively low level of expression of both Agno and VP1 during the second round of life cycle correlates our findings from the replication assays (Fig. 10A), suggesting that the mitochondrial targeting sequence of Agno appears to play an important role in successful completion of the viral replication cycle. However, since agnoprotein targets various proteins and protein complexes in cells (Saribas et al., 2020) and additional roles for the MTS sequences were also previously reported by others (Suzuki et al., 2010), thus the observed effect of Agno MTS mutant on viral replication could also be attributed to the combination of its effects exerted on the infected cells and not solely to its mitochondrial targeting function”.
4. Conclusions
Agnoprotein is one of the important regulatory proteins of JCV. In the absence of its expression, JCV is unable to sustain its productive life cycle. It is a multifunctional protein with multiple functional domains. It targets a large number of host proteins and organelles, including mitochondria and contains a mitochondrial targeting sequence located at its N-terminus. In this report, we have investigated the functional consequences of mitochondrial targeting by Agno and demonstrated its negative effects on the mitochondrial functions, including ATP production, membrane potential and respiration (Fig. 4, 6, 7 and 9). Consistent with these findings, ROS production by mitochondria was also found to be substantially increased in Agno-positive cells (Fig. 8). Another interesting finding was a substantial increase in Ca2+ influx into mitochondria in the presence of Agno (Fig. 5). Note that Ca2+ signaling plays an important role in the stimulation of the electron transport complexes and therefore ATP production. However, if the Ca2+ influx exceeds a certain threshold in mitochondria, this could then lead to the activation of mitochondrial permeability transition pore (mPTP) complex and thereby the loss of mitochondrial membrane potential and cell death (De Stefani et al., 2011). We believe that Agno plays a differential role in Ca2+ signaling in mitochondria with respect to the stimulation of Ca2+-coupled ATP production and activation of the mPTP complex during the JCV replication cycle. That is, during the early phases of the infection cycle, as the expression of Agno increases, Agno stimulates Ca2+ influx into the mitochondria and therefore the ATP production. Then the ATP pool is maximally used by the infected cells to facilitate the viral replication and virion formation. Towards the late phases of the infection cycle, however, as the agnoprotein expression further increases, the Agno-mediated Ca2+ influx into the mitochondria reaches a certain threshold at which the harmful effects of Ca2+ begin to manifest itself, such as the start of the loss of mitochondrial membrane potential and cell death, which in turn help the release of the infectious virions to initiate the next round of the infection cycle. Further studies are required to distinguish between these scenarios.
Interaction of Agno with mitochondria was clearly demonstrated by mitochondria purifications (Fig. 2B) and the mitochondrial membrane fractionation experiments (Fig. 3A), but Agno lacking MTS signal was still found to be co-fractionated with mitochondria fractions (Fig 2B and 3B) suggesting that additional regions within Agno is also involved in this targeting process. We believe that the major α-helical region of Agno most likely also plays a role in this targeting, because this region was previously determined to mediate not only homo-dimerization/–oligomerization but also interaction with various cellular and viral proteins (Saribas et al., 2019). In a support such an assumption, Manzetti et al., recently reported that the dimerization domain of BKV Agno protein contributes to its mitochondrial targeting (Manzetti et al., 2020). In addition, an MTS deletion mutant virus was used to assess the impact of MTS on viral replication. Results clearly showed a significant reduction in the replication efficiency of the mutant compared to WT (Fig. 10A and B) indicating the importance of the MTS signal in Agno function during the viral replication cycle.
Mitochondrial targeting by Agno was also supported by our recent proteomic studies, which revealed the interaction of Agno with a substantial number of mitochondrial proteins (nearly 50), some of which are the structural components of the mitochondrial protein import pathways, electron transport chain (ETC) complexes and metabolic enzymes (Saribas et al., 2020). Regarding the mitochondrial protein import pathways, we found the interaction of Agno with components of the outer (TOM) and inner (TIM) membrane translocase complexes, which are essential for the mitochondrial maturation and survival (Becker et al., 2012; Harbauer et al., 2014; Rehling et al., 2004). Particularly, its interaction with TOM70 and TOM22 in the TOM complex and with several components of TIM23 complex suggests that Agno may dysregulate the mitochondrial functions by targeting its protein import pathways. However, in mitochondrial co-fractionation studies, we observed the interaction of Agno only with the outer membrane components. This apparently shows a discrepancy with our findings from the proteomic studies. Such a discrepancy can be explained by the use of the different buffer systems in those experiments and therefore exhibition of a differential stability of the interaction of Agno with its target proteins. In sum, our recent proteomic studies provided us a road map to analyze the effects of Agno on various pathways and organelles. In this report, we assessed only the global effect of Agno on mitochondrial functions but still so many mechanistic questions remained unanswered such as how Agno induces Ca2+ influx into mitochondria and what are the molecular mechanisms for this process?, does JCV Agno alter mitochondria-mediated innate and adoptive immunity? (Manzetti et al., 2020; Weinberg et al., 2015), what are the mechanisms of the targeting of the electron transport system and metabolic pathways by Agno? We just started understanding of the remarkable functions of Agno during JCV life cycle.
Supplementary Material
Research highlights:
Agno is one of the critical regulatory proteins of JC virus and plays important roles during the JC virus life cycle.
Agno targets mitochondria through its MTS and dimerization domain; and negatively affects mitochondrial functions.
Replication efficiency of JCV is significantly reduced when the Agno MTS is deleted.
Acknowledgments
This work was supported by a NIH grant (R01NS090949.) awarded to MS. The primary human fetal astrocyte (PHFA) cells were provided by the core facility of the Comprehensive NeuroAIDS Center (CNAC, Temple University) funded by a NIH grant (P30MH09177). DT and PJ are supported by NIH pathway to independence awards, K99DK120876 and K99AG065445 respectively. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version of the publication.
Conflict of interest
Authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afanasyeva MA, Ustiugova AS, Golyshev SA, Kopylov AT, Bogolyubova AV, Demin DE, Belousov PV, Schwartz AM, 2018. Isolation of Large Amounts of Highly Pure Mitochondria for “Omics” Studies. Biochemistry-Moscow+ 83, 76–85. [DOI] [PubMed] [Google Scholar]
- Anand SK, Tikoo SK, 2013. Viruses as modulators of mitochondrial functions. Adv Virol 2013, 738794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma A, Matsuo A, Suzuki T, Kurosawa T, Zhang X, Aida Y, 2006. Human immunodeficiency virus type 1 Vpr induces cell cycle arrest at the G(1) phase and apoptosis via disruption of mitochondrial function in rodent cells. Microbes Infect 8, 670–679. [DOI] [PubMed] [Google Scholar]
- Baum MK, Sales S, Jayaweera DT, Lai S, Bradwin G, Rafie C, Page JB, Campa A, 2011. Coinfection with hepatitis C virus, oxidative stress and antioxidant status in HIV-positive drug users in Miami. HIV Med 12, 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Bottinger L, Pfanner N, 2012. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci 37, 85–91. [DOI] [PubMed] [Google Scholar]
- Berger JR, 2000. Progressive Multifocal Leukoencephalopathy. Curr Treat Options Neurol 2, 361–368. [DOI] [PubMed] [Google Scholar]
- Berger JR, Kaszovitz B, Post MJ, and Dickinson G., 1987. Progressive multifocal leukoencephalopathy associated with immunodeficiency virus infection. A review of the literature with a report of sixteen cases. Ann Intern Med. 107, 78–87. [DOI] [PubMed] [Google Scholar]
- Bernardi P, 1999. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol Rev 79, 1127–1155. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Lipp P, 1998. Calcium--a life and death signal. Nature 395, 645–648. [DOI] [PubMed] [Google Scholar]
- Boya P, Pauleau A-L, Poncet D, Gonzalez-Polo R-A, Zamzami N, Kroemer G, 2004. Viral proteins targeting mitochondria: controlling cell death. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1659, 178–189. [DOI] [PubMed] [Google Scholar]
- Boya P, Roumier T, Andreau K, Gonzalez-Polo RA, Zamzami N, Castedo M, Kroemer G, 2003. Mitochondrion-targeted apoptosis regulators of viral origin. Biochem Bioph Res Co 304, 575–581. [DOI] [PubMed] [Google Scholar]
- Boyman L, Karbowski M, Lederer WJ, 2019. Regulation of Mitochondrial ATP Production: Ca(2+) Signaling and Quality Control. Trends Mol Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisac C, Teoule F, Autret A, Pelletier I, Colbere-Garapin F, Brenner C, Lemaire C, Blondel B, 2010. Calcium flux between the endoplasmic reticulum and mitochondrion contributes to poliovirus-induced apoptosis. J Virol 84, 12226–12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito AF, Pinney JW, 2017. Protein-Protein Interactions in Virus–Host Systems. Front Microbiol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW, 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7, 1306–1312. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P, 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal of Biochemistry 241, 779–786. [DOI] [PubMed] [Google Scholar]
- Cloonan SM, Choi AM, 2012. Mitochondria: commanders of innate immunity and disease? Curr Opin Immunol 24, 32–40. [DOI] [PubMed] [Google Scholar]
- Coric P, Saribas AS, Abou-Gharbia M, Childers W, Condra JH, White MK, Safak M, Bouaziz S, 2017. Nuclear Magnetic Resonance Structure of the Human Polyoma JC Virus Agnoprotein. J Cell Biochem 118, 3268–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric P, Saribas AS, Abou-Gharbia M, Childers W, White MK, Bouaziz S, Safak M, 2014. Nuclear magnetic resonance structure revealed that the human polyomavirus JC virus agnoprotein contains an alpha-helix encompassing the Leu/Ile/Phe-rich domain. J Virol 88, 6556–6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie M, Cicalese S, Sariyer IK, 2018. Neuroimmune Regulation of JC Virus by Intracellular and Extracellular Agnoprotein. J Neuroimmune Pharmacol 13, 126–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la Fuente Perez S, Bogorad R, Koteliansky V, Adijanto J, Mootha VK, Hajnoczky G, 2013. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)(+) uniporter. Cell Metab 17, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X, Vidal JE, Oliveira A.C.P.d., Simpson DM, Morgello S, Hecht JH, Ngo LH, Koralnik IJ, 2012. JC virus granule cell neuronopathy is associated with VP1 C terminus mutants. J Gen Virol 93, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinyan A, Darbinian N, Safak M, Radhakrishnan S, Giordano A, Khalili K, 2002. Evidence for dysregulation of cell cycle by human polyomavirus, JCV, late auxiliary protein. Oncogene 21, 5574–5581. [DOI] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R, 2011. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle L, Gordon J, Enam S, Delbue S, Croul S, Abraham S, Radhakrishnan S, Assimakopoulou M, Katsetos CD, Khalili K, 2002. Expression of human neurotropic polyomavirus JCV late gene product agnoprotein in human medulloblastoma. J Natl Cancer Inst 94, 267–273. [DOI] [PubMed] [Google Scholar]
- Doonan PJ, Chandramoorthy HC, Hoffman NE, Zhang XQ, Cardenas C, Shanmughapriya S, Rajan S, Vallem S, Chen XW, Foskett JK, Cheung JY, Houser SR, Madesh M, 2014. LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation. Faseb Journal 28, 4936–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan PJ, Chandramoorthy HC, Hoffman NE, Zhang XQ, Cardenas C, Shanmughapriya S, Rajan S, Vallem S, Chen XW, Foskett JK, Cheung JY, Houser SR, Madesh M, 2015. LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation (vol 28, pg 4936, 2014). Faseb Journal 29, 3593–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Pasquier RA, Corey S, Margolin DH, Williams K, Pfister LA, De Girolami U, Mac Key JJ, Wuthrich C, Joseph JT, Koralnik IJ, 2003. Productive infection of cerebellar granule cell neurons by JC virus in an HIV+ individual. Neurology 61, 775–782. [DOI] [PubMed] [Google Scholar]
- Dugan AS, Gasparovic ML, Tsomaia N, Mierke DF, O’Hara BA, Manley K, Atwood WJ, 2007. Identification of amino acid residues in BK virus VP1 that are critical for viability and growth. J Virol 81, 11798–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE, 2005. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6, 197–208. [DOI] [PubMed] [Google Scholar]
- Ellis LC, Koralnik IJ, 2015. JC virus nucleotides 376-396 are critical for VP1 capsid protein expression. J Neurovirol 21, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczy MW, Marshall LJ, Nelson CD, Atwood WJ, Nath A, Khalili K, Major EO, 2012. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev 25, 471–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AL, 2005. Natively unfolded proteins. Curr Opin Struct Biol 15, 35–41. [DOI] [PubMed] [Google Scholar]
- Foy E, Li K, Sumpter R Jr., Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M Jr., 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A 102, 2986–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko A, Baris OR, Bergschneider E, von Toerne C, Hauck SM, Aichler M, Walch AK, Wurst W, Wiesner RJ, Johnston ICD, de Angelis MH, 2013. Efficient Isolation of Pure and Functional Mitochondria from Mouse Tissues Using Automated Tissue Disruption and Enrichment with Anti-TOM22 Magnetic Beads. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Xia Y, 2011. Structural principles within the human-virus protein-protein interaction network. Proceedings of the National Academy of Sciences 108, 10538–10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque RJ, Bream GL, Cannella MT, 1984. Human polyomavirus JC virus genome. J Virol 51, 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits N, Moens U, 2012. Agnoprotein of mammalian polyomaviruses. Virology 432, 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW, 2003. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol 77, 7214–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, Carrasco L, 2003. Viroporins. FEBS Lett 552, 28–34. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ., 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52, 456–467. [DOI] [PubMed] [Google Scholar]
- Granatiero V, De Stefani D, Rizzuto R, 2017. Mitochondrial Calcium Handling in Physiology and Disease. Adv Exp Med Biol 982, 25–47. [DOI] [PubMed] [Google Scholar]
- Griffin SD, Harvey R, Clarke DS, Barclay WS, Harris M, Rowlands DJ, 2004. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J Gen Virol 85, 451–461. [DOI] [PubMed] [Google Scholar]
- Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C, 2014. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab 19, 357–372. [DOI] [PubMed] [Google Scholar]
- Hu L, Chen L, Yang G, Li L, Sun H, Chang Y, Tu Q, Wu M, Wang H, 2011. HBx sensitizes cells to oxidative stress-induced apoptosis by accelerating the loss of Mcl-1 protein via caspase-3 cascade. Mol Cancer 10, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacotot E, Ravagnan L, Loeffler M, Ferri KF, Vieira HL, Zamzami N, Costantini P, Druillennec S, Hoebeke J, Briand JP, Irinopoulou T, Daugas E, Susin SA, Cointe D, Xie ZH, Reed JC, Roques BP, Kroemer G, 2000. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J Exp Med 191, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt-DeMasters BK, Tyler KL, 2005. Progressive multifocal leukoencephalopathy complicating treatment with natalizumab and interferon beta-1a for multiple sclerosis. N Engl J Med 353, 369–374. [DOI] [PubMed] [Google Scholar]
- Lambert JP, Luongo TS, Tomar D, Jadiya P, Gao E, Zhang X, Lucchese AM, Kolmetzky DW, Shah NS, Elrod JW, 2019. MCUB Regulates the Molecular Composition of the Mitochondrial Calcium Uniporter Channel to Limit Mitochondrial Calcium Overload During Stress. Circulation 140, 1720–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer-Gould A, Atlas SW, Green AJ, Bollen AW, Pelletier D, 2005. Progressive multifocal leukoencephalopathy in a patient treated with natalizumab. N Engl J Med 353, 375–381. [DOI] [PubMed] [Google Scholar]
- Lassoued S, Gargouri B, El Feki Ael F, Attia H, Van Pelt J, 2010. Transcription of the Epstein-Barr virus lytic cycle activator BZLF-1 during oxidative stress induction. Biol Trace Elem Res 137, 13–22. [DOI] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr., Lemon SM, 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 102, 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Boehning DF, Qian T, Popov VL, Weinman SA, 2007. Hepatitis C virus core protein increases mitochondrial ROS production by stimulation of Ca2+ uniporter activity. FASEB J 21, 2474–2485. [DOI] [PubMed] [Google Scholar]
- Lund K, Ziola B, 1985. Cell sonicates used in the analysis of how measles and herpes simplex type 1 virus infections influence Vero cell mitochondrial calcium uptake. Can J Biochem Cell Biol 63, 1194–1197. [DOI] [PubMed] [Google Scholar]
- Macho A, Calzado MA, Jimenez-Reina L, Ceballos E, Leon J, Munoz E, 1999. Susceptibility of HIV-1-TAT transfected cells to undergo apoptosis. Biochemical mechanisms. Oncogene 18, 7543–7551. [DOI] [PubMed] [Google Scholar]
- Madesh M, Hawkins BJ, Milovanova T, Bhanumathy CD, Joseph SK, Ramachandrarao SP, Sharma K, Kurosaki T, Fisher AB, 2005. Selective role for superoxide in InsP3 receptor-mediated mitochondrial dysfunction and endothelial apoptosis. J Cell Biol 170, 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J, 1985. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci U S A 82, 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Cardenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenar T, Csordas G, Madireddi P, Yang J, Muller M, Miller R, Kolesar JE, Molgo J, Kaufman B, Hajnoczky G, Foskett JK, Madesh M, 2012a. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nature Cell Biology 14, 1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Doonan P, Cardenas C, Chandramoorthy HC, Muller M, Miller R, Hoffman NE, Gandhirajan RK, Molgo J, Birnbaum MJ, Rothberg BS, Mak DO, Foskett JK, Madesh M, 2012b. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell 151, 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzetti J, Weissbach FH, Graf FE, Unterstab G, Wernli M, Hopfer H, Drachenberg CB, Rinaldo CH, Hirsch HH, 2020. BK Polyomavirus Evades Innate Immune Sensing by Disrupting the Mitochondrial Network and Promotes Mitophagy. Iscience 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi S, Bittremieux M, Missiroli S, Morganti C, Patergnani S, Sbano L, Rimessi A, Kerkhofs M, Parys JB, Bultynck G, Giorgi C, Pinton P, 2017. Endoplasmic Reticulum-Mitochondria Communication Through Ca(2+) Signaling: The Importance of Mitochondria-Associated Membranes (MAMs). Adv Exp Med Biol 997, 49–67. [DOI] [PubMed] [Google Scholar]
- Merabova N, Kaniowska D, Kaminski R, Deshmane SL, White MK, Amini S, Darbinyan A, Khalili K, 2008. JC virus agnoprotein inhibits in vitro differentiation of oligodendrocytes and promotes apoptosis. J Virol 82, 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco MC, Jensen PN, Hou J, Durham LC, Major EO, 1998. Detection of JC virus DNA in human tonsil tissue: evidence for site of initial viral infection. J Virol 72, 9918–9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Love J, Gee GV, O’Hara BA, Assetta B, Atkinson AL, Dugan AS, Haley SA, Atwood WJ, 2019. JC Polyomavirus Uses Extracellular Vesicles To Infect Target Cells. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura S, Khoury G, Jay G, 1983. Subcellular localization of the simian virus 40 agnoprotein. J Virol 45, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara BA, Gee GV, Atwood WJ, Haley SA, 2018. Susceptibility of Primary Human Choroid Plexus Epithelial Cells and Meningeal Cells to Infection by JC Virus. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara BA, Morris-Love J, Gee GV, Haley SA, Atwood WJ, 2020. JC Virus infected choroid plexus epithelial cells produce extracellular vesicles that infect glial cells independently of the virus attachment receptor. PLoS Pathog 16, e1008371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Endo S, Takahashi H, Sawa H, Umemura T, Nagashima K, 2001. Distribution and function of JCV agnoprotein. J Neurovirol 7, 302–306. [DOI] [PubMed] [Google Scholar]
- Otlu O, De Simone FI, Otalora YL, Khalili K, Sariyer IK, 2014. The agnoprotein of polyomavirus JC is released by infected cells: evidence for Its cellular uptake by uninfected neighboring cells. Virology 468-470, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Kwiatkowski W, Deerinck TJ, Noske A, Blain KY, Land HS, Soria C, Powers CJ, May AP, Shu X, Tsien RY, Fitzpatrick JA, Long JA, Ellisman MH, Choe S, O’Shea CC, 2012. A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors. Cell 151, 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett BL, Zu Rhein GM, Walker DL, Echroade R, and Dessel B., 1971. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet. i, 1257–1260. [DOI] [PubMed] [Google Scholar]
- Patron M, Raffaello A, Granatiero V, Tosatto A, Merli G, De Stefani D, Wright L, Pallafacchina G, Terrin A, Mammucari C, Rizzuto R, 2013. The mitochondrial calcium uniporter (MCU): molecular identity and physiological roles. J Biol Chem 288, 10750–10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK, 2010. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature 467, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, 2000. Protein sorting: recognizing mitochondrial presequences. Curr Biol 10, R412–415. [DOI] [PubMed] [Google Scholar]
- Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li AA, Girgis HS, Kuchimanchi S, De Groot J, Speciner L, Taneja N, Oshea J, Koteliansky V, Mootha VK, 2013. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One 8, e55785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poenisch M, Burger N, Staeheli P, Bauer G, Schneider U, 2009. Protein X of Borna disease virus inhibits apoptosis and promotes viral persistence in the central nervous systems of newborn-infected rats. J Virol 83, 4297–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo CW, Yang YL, Yoo NK, Choi SY, 2008. Reactive oxygen species activate HIV long terminal repeat via post-translational control of NF-kappaB. Biochem Biophys Res Commun 376, 180–185. [DOI] [PubMed] [Google Scholar]
- Raffaello A, De Stefani D, Sabbadin D, Teardo E, Merli G, Picard A, Checchetto V, Moro S, Szabo I, Rizzuto R, 2013. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. Embo J 32, 2362–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N, 2004. Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol 5, 519–530. [DOI] [PubMed] [Google Scholar]
- Rinaldo CH, Hirsch HH, 2007. Antivirals for the treatment of polyomavirus BK replication. Expert Rev Anti Infect Ther 5, 105–115. [DOI] [PubMed] [Google Scholar]
- Rinaldo CH, Traavik T, Hey A, 1998. The agnogene of the human polyomavirus BK is expressed. J Virol 72, 6233–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S, Frye RE, Slattery J, Wynne R, Tippett M, Pavliv O, Melnyk S, James SJ, 2014. Oxidative stress induces mitochondrial dysfunction in a subset of autism lymphoblastoid cell lines in a well-matched case control cohort. PLoS One 9, e85436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K, 2001. Interaction of JC Virus Agno Protein with T Antigen Modulates Transcription and Replication of the Viral Genome in Glial Cells. J Virol 75, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovacs-Bogdan E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, Calvo SE, Goldberger O, Mootha VK, 2013. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342, 1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas A, Abou-Gharbia M, Childers W, Sariyer IK, White MK, Safak M, 2013. Essential roles of Leu/Ile/Phe-rich domain of JC virus agnoprotein in dimer/oligomer formation, protein stability and splicing of viral transcripts. Virology 443, 161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Arachea BT, White MK, Viola RE, Safak M, 2011. Human polyomavirus JC small regulatory agnoprotein forms highly stable dimers and oligomers: implications for their roles in agnoprotein function. Virology 420, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Coric P, Bouaziz S, Safak M, 2019. Expression of novel proteins by polyomaviruses and recent advances in the structural and functional features of agnoprotein of JC virus, BK virus, and simian virus 40. J Cell Physiol 234, 8295–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Coric P, Hamazaspyan A, Davis W, Axman R, White MK, Abou-Gharbia M, Childers W, Condra JH, Bouaziz S, Safak M, 2016. Emerging From the Unknown: Structural and Functional Features of Agnoprotein of Polyomaviruses. J Cell Physiol 231, 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Datta PK, Safak M, 2020. A comprehensive proteomics analysis of JC virus Agnoprotein-interacting proteins: Agnoprotein primarily targets the host proteins with coiled-coil motifs. Virology 540, 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, DeVoto J, Golla A, Wollebo HS, White MK, Safak M, 2018a. Discovery and characterization of novel trans-spliced products of human polyoma JC virus late transcripts from PML patients. J Cell Physiol 233, 4137–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, White MK, Safak M, 2012. JC virus agnoprotein enhances large T antigen binding to the origin of viral DNA replication: evidence for its involvement in viral DNA replication. Virology 433, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, White MK, Safak M, 2018b. Structure-based release analysis of the JC virus agnoprotein regions: A role for the hydrophilic surface of the major alpha helix domain in release. J Cell Physiol 233, 2343–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Akan I, Palermo V, Gordon J, Khalili K, Safak M, 2006. Phosphorylation mutants of JC virus agnoprotein are unable to sustain the viral infection cycle. J Virol 80, 3893–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Saribas AS, White MK, Safak M, 2011. Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virol J 8, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Sjoling S, Wallin E, Wrede P, Glaser E, von Heijne G, 1998. Feature-extraction from endopeptidase cleavage sites in mitochondrial targeting peptides. Proteins 30, 49–60. [PubMed] [Google Scholar]
- Sharon-Friling R, Goodhouse J, Colberg-Poley AM, Shenk T, 2006. Human cytomegalovirus pUL37×1 induces the release of endoplasmic reticulum calcium stores. Proc Natl Acad Sci U S A 103, 19117–19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani-Meigooni DN, Schwetye KE, Angeles MR, Ryschkewitsch CF, Major EO, Dang X, Koralnik IJ, Schmidt RE, Clifford DB, Kuhlmann FM, Bucelli RC, 2017. JC virus granule cell neuronopathy in the setting of chronic lymphopenia treated with recombinant interleukin-7. J Neurovirol 23, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Makino Y, Okada Y, Sunden Y, Hasegawa H, Hall WW, Sawa H, 2013. Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc Natl Acad Sci U S A 110, 18668–18673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, Nagashima K, Hall WW, Sawa H, 2010. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog 6, e1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Semba S, Sunden Y, Orba Y, Kobayashi S, Nagashima K, Kimura T, Hasegawa H, Sawa H, 2012. Role of JC virus agnoprotein in virion formation. Microbiol Immunol 56, 639–646. [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R, 2006. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol 175, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson BJ, 2001. Viruses and apoptosis. Int J Exp Pathol 82, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar D, Dong Z, Shanmughapriya S, Koch DA, Thomas T, Hoffman NE, Timbalia SA, Goldman SJ, Breves SL, Corbally DP, Nemani N, Fairweather JP, Cutri AR, Zhang X, Song J, Jana F, Huang J, Barrero C, Rabinowitz JE, Luongo TS, Schumacher SM, Rockman ME, Dietrich A, Merali S, Caplan J, Stathopulos P, Ahima RS, Cheung JY, Houser SR, Koch WJ, Patel V, Gohil VM, Elrod JW, Rajan S, Madesh M, 2016. MCUR1 Is a Scaffold Factor for the MCU Complex Function and Promotes Mitochondrial Bioenergetics. Cell Rep 15, 1673–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterstab G, Gosert R, Leuenberger D, Lorentz P, Rinaldo CH, Hirsch HH, 2010. The polyomavirus BK agnoprotein co-localizes with lipid droplets. Virology 399, 322–331. [DOI] [PubMed] [Google Scholar]
- Van Assche G, Van Ranst M, Sciot R, Dubois B, Vermeire S, Noman M, Verbeeck J, Geboes K, Robberecht W, Rutgeerts P, 2005. Progressive multifocal leukoencephalopathy after natalizumab therapy for Crohn’s disease. N Engl J Med 353, 362–368. [DOI] [PubMed] [Google Scholar]
- Van Petegem F, 2012. Ryanodine receptors: structure and function. J Biol Chem 287, 31624–31632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G, 1986. Mitochondrial targeting sequences may form amphiphilic helices. Embo J 5, 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Sena LA, Chandel NS, 2015. Mitochondria in the regulation of innate and adaptive immunity. Immunity 42, 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White FA 3rd, Ishaq M, Stoner GL, Frisque RJ, 1992. JC virus DNA is present in many human brain samples from patients without progressive multifocal leukoencephalopathy. J Virol 66, 5726–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, 2017. Mitochondrial Machineries for Protein Import and Assembly. Annu Rev Biochem 86, 685–714. [DOI] [PubMed] [Google Scholar]
- Wuthrich C, Batson S, Anderson MP, White LR, Koralnik IJ, 2016. JC Virus Infects Neurons and Glial Cells in the Hippocampus. J Neuropathol Exp Neurol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuthrich C, Cheng YM, Joseph JT, Kesari S, Beckwith C, Stopa E, Bell JE, Koralnik IJ, 2009. Frequent infection of cerebellar granule cell neurons by polyomavirus JC in progressive multifocal leukoencephalopathy. J Neuropathol Exp Neurol 68, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorova LD, Popkov VA, Plotnikov EY, Silachev DN, Pevzner IB, Jankauskas SS, Babenko VA, Zorov SD, Balakireva AV, Juhaszova M, Sollott SJ, Zorov DB, 2018. Mitochondrial membrane potential. Anal Biochem 552, 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


