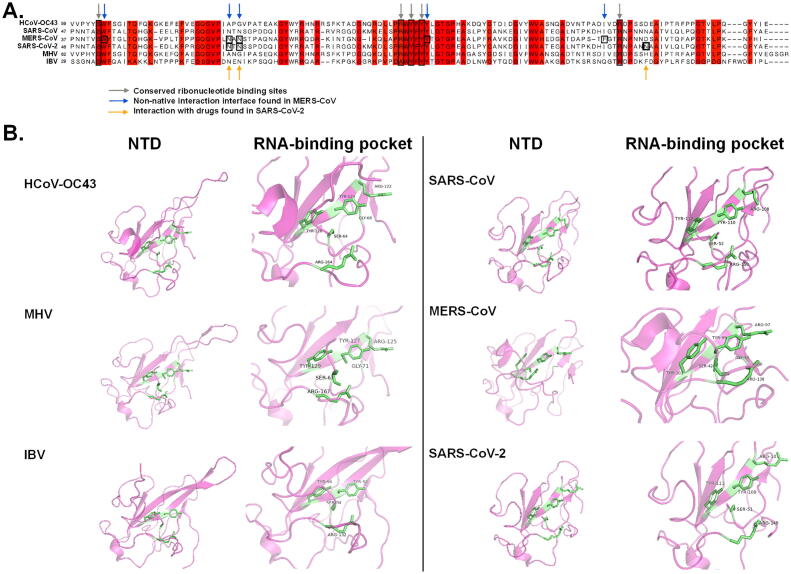
Fig. 1.
Sequence alignment and structure analyses of NTD of CoV NP. (A) Multiple sequence alignment of HCoV-OC43 (NC005147), SARS-CoV (NC004718), MERS-CoV (NC019843), SARS-CoV-2 (NC045512), MHV (NC001846) and IBV (AY692454). The highly conserved residues were highlighted in red. Grey arrows indicate conserved RNA binding sites identified in previous reports [39], [48], [51], [61]. Blue arrows indicate important residues for non-native oligomerization [48]. Yellow arrows indicate binding sites between potential anti-SARS-CoV-2 compounds and N protein [51]. (B) NTD and RNA binding pockets of HCoV-OC43 (PDB: 4J3K), MHV (PDB: 3HD4), IBV (PDB: 2GEC), SARS-CoV (PDB: 2OG3), MERS-CoV (PDB: 6KL2) and SARS-CoV-2 (PDB: 6WKP) N protein. Green sticks indicate RNA-interaction residues. HCoV-OC43, human coronavirus OC43; MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MHV, mouse hepatitis virus; IBV, infectious bronchitis virus. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)