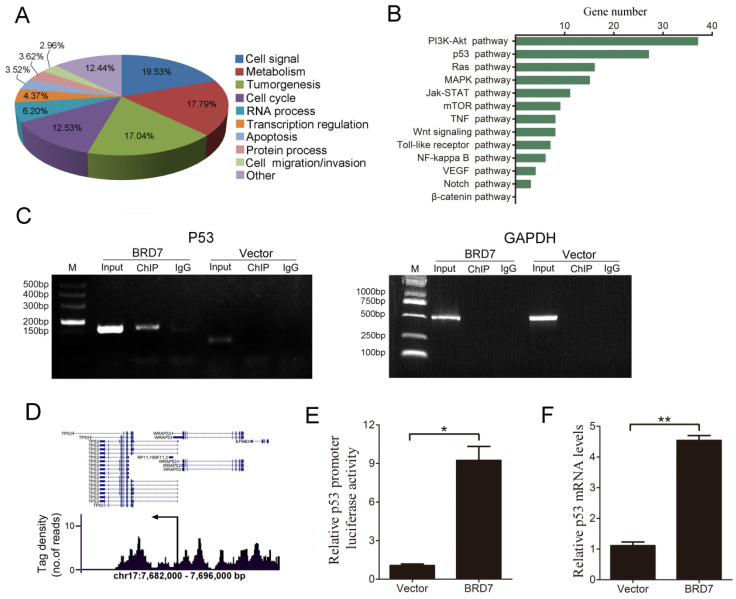
Figure 1.
Defining genome-wide BRD7 binding genes by ChIP-sequencing in Bel7402 cells transfected with BRD7. (A) Functional annotation of BRD7-associated genes based on enriched GO terms. (B) The most significant “pathways in cancer” enriched by BRD7 target genes are presented, based on KEGG pathway analysis. (C) ChIP-PCR analysis of the binding of BRD7 to p53 gene in Bel7402 cells transfected with BRD7 or Vector. Primers were designed against the peak interval sequence, and GAPDH served as the negative control. (D) Representative tag density profiles for BRD7-bound regions located in p53, and their proximity to the transcriptional initiation site (arrow). (E) The potential p53 promoter activity in Bel7402 cells transfected with BRD7 by the dual-luciferase reporter assay, normalized to the pRL-TK vector containing Renilla luciferase. (F) Fold changes in p53 mRNA levels in Bel7402 cells transfected with BRD7 compared to Bel7402 cells transfected with Vector. * p<0.05, ** p<0.01.