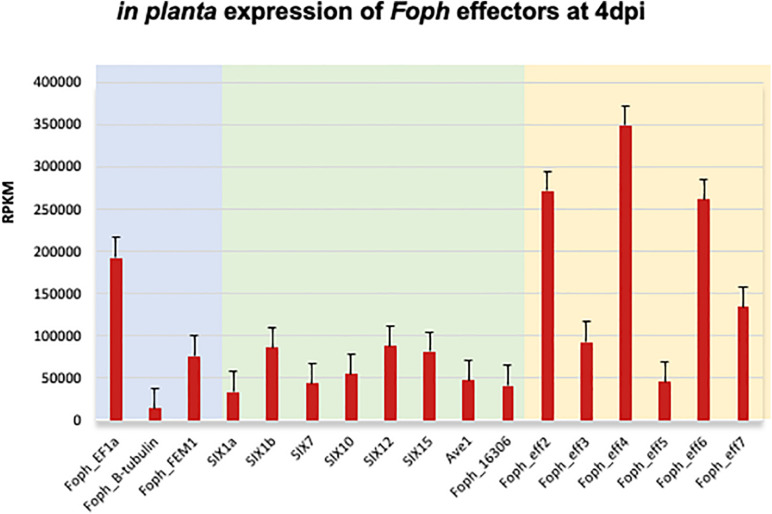
FIGURE 4.
Expression analysis of the effectors identified in the genome sequence of Foph_MAP5, using the RNAseq data form cape gooseberry susceptible plants inoculated with Foph at 4 dpi, reported in our previous in planta transcriptomic analysis. Pale blue panel indicate the genes translation elongation factor alpha (EF1a), tubulin B-chain (B-tubulin), and Fusarium Extracellular Matrix 1 (FEM1), to use as constitutive expressed control genes of Foph during host infection. Pale green indicates the expression of the homologous effectors identified in Foph. Pale yellow indicates the expression of the newly identified effectors in Foph. Six out of seven new effector candidates are expressed during cape gooseberry infection with a higher expression of eff2, eff4, and eff6, compared to the rest of the effectors analyzed. RPKM, reads per kilobase per million of mapped reads. Scale bars indicate standard error.