Figure 3.
TAP-independent peptide presentation is functional in healthy donor cells
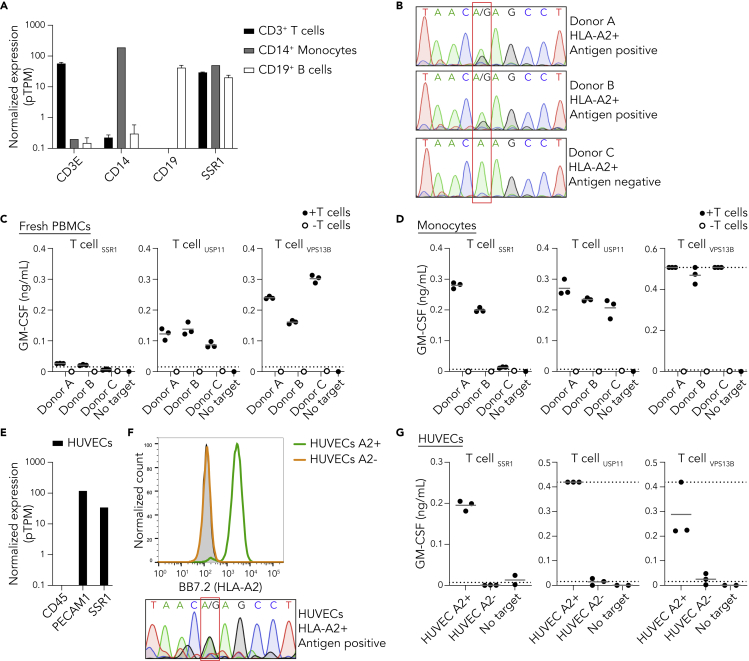
(A) Normalized expression of SSR1 and control CD14, CD19, and CD3E per depicted PBMC subset using the RNA HPA blood cell gene data (Uhlen et al., 2015). Data are represented as mean with SD.
(B) Sanger sequencing of the SSR1 SNP rs10004 of three HLA-A2 positive healthy donors, the position of the SNP is highlighted by the red box and consequence for antigen status is depicted.
(C) T cell coculture with PBMCs from three healthy donors (B), culture supernatant was analyzed by GM-CSF ELISA (IFN-γ data in Figure S4). Results from triplicate cultures are shown. Dotted lines represent the detection limits of the ELISA.
(D) As in (C) but using monocytes as target cells (IFN-γ data in Figure S4).
(E) Expression analysis of SSR1 and control CD45 and PECAM1 in TERT2 immortalized HUVECs using the RNA HPA cell line gene data.
(F) Flow cytometric analysis of HLA-A2 expression by HUVECs. HLA-A2 positive FACS sorted (green), HLA-A2 negative FACS sorted (orange) populations and unstained HUVECs (gray). Lower panel depicts rs10004 sequence of the HLA-A2 positive sorted HUVECs.
(G) T cell coculture with HLA-A2 positive and negative HUVECs (F). Culture supernatant was analyzed by GM-CSF ELISA (IFN-γ data in Figure S4). Each dot represents an individual measurement of triplicate cultures.
See also Figures S1 and S4.