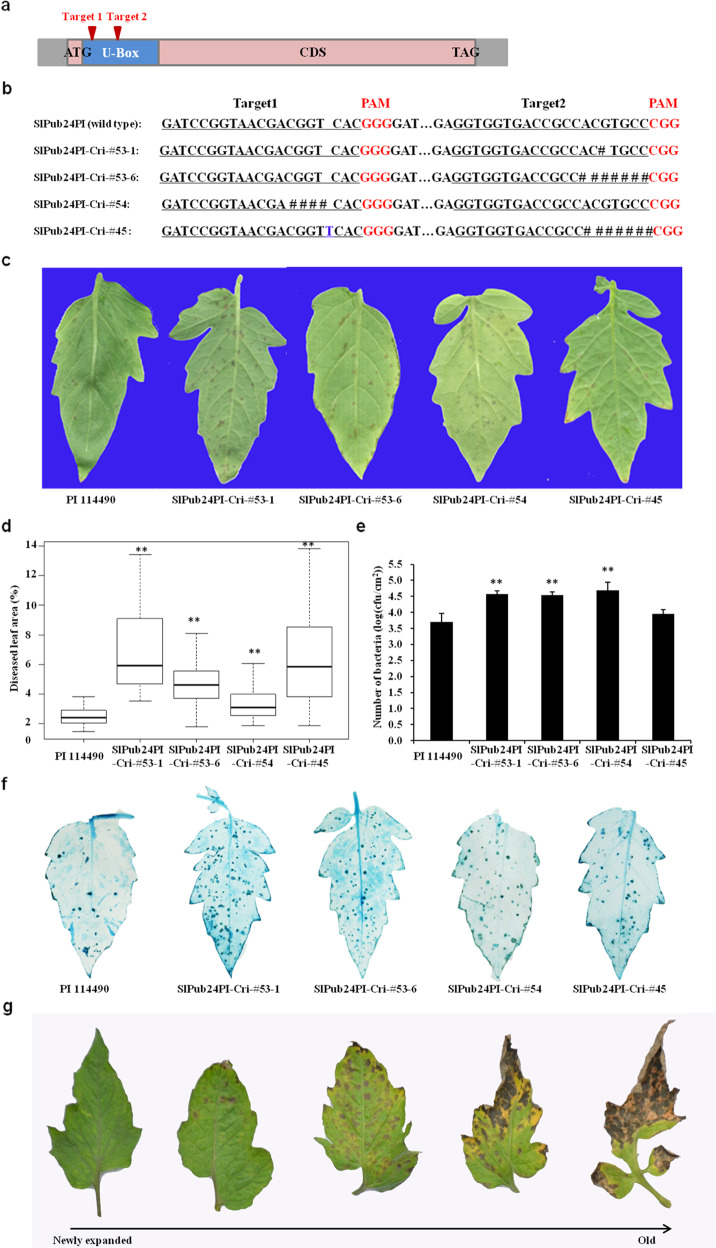
Fig. 4. Knockout of SlPub24 in the resistant line PI 114490 by the CRISPR/Cas9 editing system leads to decreased resistance to Xanthomonaseuvesicatoria pv. perforans race T3 strain Xv829 and spontaneous cell death in leaves in mutants.
a Schematic illustration of the two sgRNA target sites (red arrows) in SlPub24. b Mutations identified in four T2 mutant lines. Red font indicates protospacer-adjacent motif (PAM) sequences, and the sgRNA target sequence is underlined. c Symptoms on PI 114490 and mutant leaves at 7 days post inoculation (dpi). d Statistical analysis of disease in PI 114490 and mutants at 7 dpi. Error bars represent the SD (n = 30). e Bacterial population in leaves of PI 114490 and mutants at 9 dpi. Error bars represent the SD (n = 30). f Trypan blue staining of diseased PI 114490 and mutant leaves (n = 30). The asterisks indicate statistical significance by t test (**P < 0.01). g Spontaneous cell death on leaves of mutants at various stages, from newly expanded to old leaves