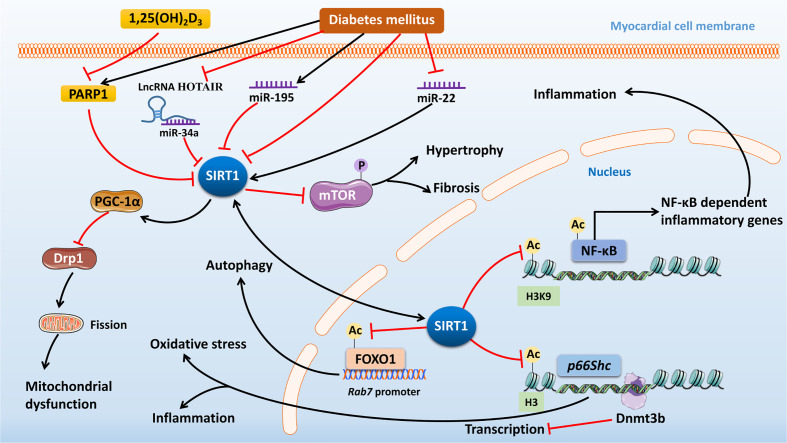
Figure 1.
The mechanisms of action and signaling pathways of SIRT1 in diabetic cardiomyopathy. Diabetes mellitus (DM) can inhibit SIRT1 expression by increased expression of PARP1, miR-34a, and miR-195, or by decreased expression of miR-22. SIRT1 can deacetylate NF-κB and H3K9 to inhibit the expression of NF-κB-dependent inflammatory genes, which improves DCM. Dysregulation of Dnmt3b and SIRT1 by DM leads to CpG demethylation and histone H3 acetylation at the p66Shc promoter, which can result in sustained transcription of p66Shc to cause cardiac oxidative stress and inflammation. SIRT1 can reduce Drp1-mediated mitochondrial fission to exert its cardioprotective role in DCM through increasing PGC-1α expression. 1,25(OH)2D3 treatment can inhibit PARP1 expression to increase the expression of SIRT1 and repress the phosphorylation of mTOR, thus improving DM-induced cardiac hypertrophy and fibrosis. And SIRT1 can deacetylate FOXO1 and enhance FOXO1 DNA binding at the Rab7 promoter region to ameliorate dysfunctional autophagic flux in the hearts of diabetic mice. Finally, lncRNA HOTAIR can upregulate SIRT1 expression by sponging miR-34a to improve DCM. SIRT1, Sirtuin 1; DM, diabetes mellitus; PARP1, poly (ADP-ribose) polymerase 1; miR, microRNA; NF-κB, nuclear factor kappa B; Dnmt3b, DNA methyltransferase 3b; DCM, diabetic cardiomyopathy; Drp1, dynamin-related protein 1; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator-1 alpha 1,25(OH)2D3, 1,25-Dihydroxyvitamin D3; mTOR, mammalian target of rapamycin; FOXO1, forkhead box O 1; lncRNA HOTAIR, long non-coding RNA HOX transcript antisense RNA.