Abstract
Although miR-101 is involved in the development and progression of T-cell acute lymphoblastic leukemia (T-ALL), the underlying molecular mechanisms remain unclear. In this article, we report that miR-101 expression was inversely correlated with CX chemokine receptor 7 (CXCR7) level in T-ALL. Introducing miR-101 inhibited T-ALL cell proliferation and invasion in vitro and suppressed tumor growth and lung metastasis in vivo. CXCR7 was identified as a direct target of miR-101. The inhibitory effects of miR-101 were mimicked and counteracted by CXCR7 depletion and overexpression, respectively. Mechanistically, miR-101 targets CXCR7/STAT3 axis to reduce T-ALL growth and metastasis. Overall, these findings implied the potential application of miR-101 and CXCR7 in T-ALL treatment.
Key words: MicroRNA-101 (miR-101), CXCR7, T-cell acute lymphoblastic leukemia (T-ALL)
INTRODUCTION
Acute lymphoblastic leukemia (ALL) is a hematologic malignancy arising from hematopoietic precursors of lymphoid cells and is the most common type of leukemia. T-cell acute lymphoblastic leukemia (T-ALL) accounts for 15% and 25% of pediatric and adult ALL cases, respectively1. Despite great improvement in T-ALL therapy, the overall survival rate is still about 60%–70% in children and only 30%–40% in adults2,3. Thus, identifying the factors and potential mechanisms contributing to T-ALL and securing advanced treatments are necessary and urgent.
MicroRNAs (miRNAs) are small noncoding RNAs that negatively regulate the gene expression by mRNA degradation or translational repression4. Alterations in miRNA expression facilitate the acquisition of cancer hallmarks5. MicroRNA-101 (miR-101) acts a putative tumor suppressor in several types of cancers, such as gastric cancer, hepatocellular carcinoma, and breast cancer6–8. miR-101 also regulates the hematological malignancies, including T-ALL9–12. Correia et al.11 revealed that downregulated miR-101 exerts inhibitory effects on T-ALL by targeting TAL1. Reportedly, miR-101 targets Notch1 to modulate T-ALL progression and chemotherapeutic sensitivity12. However, the biological functions of miR-101 in T-ALL are still unclear.
C-X-C chemokine receptor type 7 (CXCR7) is a common chemokine receptor that is highly expressed in many human cancers, such as breast cancer, glioma, non-small cell lung cancer, and prostate cancer8,13–15. The C-X-C motif ligand (CXCL) 12/CXCR7 axis regulates various oncogenic pathways and positively correlates with metastasis and poor prognosis; hence, targeting CXCR7 could be an effective strategy for cancer therapy8,16,17. Previous reports highlighted the essential role of CXCL12/CXCR7 in acute myeloid leukemia and chronic myelogenous leukemia18,19. Moreover, CXCR7 contributed to T-ALL cell migration in response to CXCL12 induction18. Nevertheless, the function and regulatory mechanism of CXCR7 in T-ALL remain unknown.
In this study, we investigated the expression and function link between miR-101 and CXCR7 and their biological roles in T-ALL. Inverse correlation between miR-101 and CXCR7 was identified in T-ALL cells and patient samples. We demonstrated that miR-101 reduced T-ALL cell proliferation and invasion in vitro by directly targeting CXCR7. miR-101 overexpression or CXCR7 depletion impaired T-ALL growth and lung metastasis in vivo. Molecular mechanism analyses involved in these processes revealed that miR-101 reduced T-ALL tumorigenesis by targeting CXCR7/signal transducer and activator of transcription 3 (STAT3) signaling pathway. Overall, the results suggest that miR-101/CXCR7/STAT3 axis is a promising therapeutic target for T-ALL.
MATERIALS AND METHODS
Patients
Healthy donors (n = 10) and patients with myelodysplastic syndrome (MDS; n = 30), acute myeloid leukemia (AML; n = 20), and acute lymphoblastic leukemia (ALL; n = 30) diagnosed in the Department of Hematology of the 150th Central Hospital of PLA were enrolled in this study. The patients were diagnosed based on routine morphological evaluation, immunophenotyping, and cytochemical smears according to the WHO criteria. The inclusion criteria for each patient were as follows: attended the clinic between 2008 and 2013; confirmed diagnosis of MDS, AML, or ALL; and untreated at the time of the study. All healthy controls and patients provided informed written consent. This study was approved by the ethics committee of the 150th Central Hospital of PLA.
Bone Marrow (BM) Sample Collection and Isolation of T Cells From BM
BM samples were harvested for mononuclear cells isolation from all the patients and healthy donors using Ficoll-Paque PLUS (Amersham Biosciences, Uppsala, Sweden) density gradient centrifugation. Normal T cells were isolated from mononuclear cells of the healthy donors by MACS depletion (Miltenyi Biotec, Bergisch Gladbach, Germany). The use of this bone marrow collection was approved by the ethics committee of the 150th Central Hospital of PLA.
Cell Culture
Human T-ALL cell lines (HPB-ALL, TALL-1, KOPTK1, Jurkat, CCRF-CEM, and Molt 4) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Molt 4-luciferase (Molt 4-luc) cells with stable luciferase expression were obtained from PerkinElmer (Santa Clara, CA, USA). T-ALL cells were cultured in Roswell Park Memorial Institute-1640 (RPMI-1640; Gibco, BRL, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco). All cells were incubated in a humidified incubator with 5% CO2 at 37°C. Cells from passages 2–4 were used for experiments.
Cell Stimulation
Cell stimulation was performed as described previously8. In brief, cells were serum starved for 4 h at 37°C. Serum-starved cells were stimulated with 100 ng/ml CXCL12 and incubated at 37°C for various time periods. At the end of the stimulation, cells were harvested for analysis.
Cell Transfection and Infection
miR-101 mimic and its negative control RNA (miR-NC) were synthesized by Qiagen (Hilden, Germany). Plasmid pcDNA3.1-CXCR7 was obtained from Biovector (Beijing, P.R. China). Transfection was performed by electroporation using Amaxa Nucleofector™ Device (Lonza, Germany) following the manufacturer’s instructions. Molt 4 cells were transfected with 100 nM miR-101 mimics or miR-NC or cotransfected with 100 nM miR-101 mimics and 2 μg of CXCR7-expressing plasmid in six-well plates for in vitro experiments. Lentivirus pGCsilence (pGCsi), pGCsi-miR-101, pLKO.1, and pLKO.1-shCXCR7 were procured from GenePharma (Shanghai, P.R. China) and infected with Molt 4-luc cells. All constructs were confirmed by DNA sequencing. After 15 days of screening with puromycin (5 μg/ml; Sigma-Aldrich, St. Louis, MO, USA), stable clones were generated and harvested for in vivo assays.
Luciferase Reporter Assay
For dual-luciferase reporter assay, the wild-type (WT) 3′-UTR of CXCR7 and a variant containing mutations in the putative miR-101 binding sites were inserted downstream of the firefly luciferase gene in the pGL3 vector (Promega, Madison, WI, USA). The primers used to amplify the WT and mutant (MUT) 3′-UTRs are as follows: WT 3′-UTR: 5′-GAATTCGACGGGTTTACTTGTTTT-3′ (forward) and 5′-CTGCAGGGAAACAAAAATCTTTAT-3′ (reverse) and MUT 3’-UTR: 5′-CACCAATAGTGAGAAATATTTCACTTAAAATTTAC-3′ (forward) and 5′-GTATTTAAATTTTAAGTGAAATATTTCTCACTATTG-3′ (reverse). All constructs were confirmed by DNA sequencing. Molt 4 cells were cotransfected with reporter constructs, an internal control vector (pGL3), and a synthetic miR-101 mimic. After 48 h of transfection, luciferase activity, normalized to the activity of Renilla, was determined using the Dual-Luciferase Reporter Assay System (Promega) and a luminometer (Glomax 20/20; Promega) following the manufacturer’s protocol.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
Total RNA was extracted from human BM samples, the cells, and tumor tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. To quantify the miR-101 expression, complementary DNA (cDNA) was synthesized with miScript Reverse Transcription kit (Qiagen) and then amplified using SYBR Premix Ex Taq™ (TaKaRa, Otsu, Shiga, Japan). U6 was used as an internal control. To quantify the mRNA levels of CXCR7, cDNA was generated using the Reverse Transcription kit (Promega). qPCR assay was performed using a standard IQTM SYBR Green Supermix kit (Bio-Rad, Berkeley, CA, USA), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The primer pairs were miR-101: 5′-CTACAGTACTGTGATAACTGAA-3′; U6: 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAAT TTGCGT-3′ (reverse), CXCR7: 5′-AGCCTGGCAACTACTCTGACA-3′ (forward) and 5′-GAAGCAC GTTCTTGTTAGGCA-3′ (reverse), and GAPDH: 5′-ACACCCACTCCTCCACCTTT-3′ (forward) and 5′-TTACTCCTTGGAGGCCATGT-3′ (reverse). Gene expression was measured in triplicate, quantified using the 2−ΔΔCT method, and normalized to a control. All qPCR assays were performed on an Applied Biosystems 7500 system (Applied Biosystems, Warrington, UK).
Cell Viability Assay
Cell viability was measured using Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan). Molt 4 cells were seeded into 96-well plates at a density of 1 × 103 cells per well (n = 5 for each time point) in a final volume of 100 μl. The cells were cultured for 1, 2, 3, or 4 days after transfection with miR-NC, miR-101 mimics, or miR-101 mimics + CXCR7-expressing plasmid. In accordance with the manufacturer’s instructions, CCK-8 solution (10 μl) was added to each well. The absorbance was measured at 450 nm after incubation for 4 h to calculate the number of viable cells.
Cell Cycle Analysis
Molt 4 cells were seeded into 12-well plates. At 48 h after transfection with RNA oligonucleotides, the cells were harvested, resuspended in PBS, fixed with ice-cold 70% ethanol, and treated with 1 mg/ml RNase at 4°C overnight. Intracellular DNA was labeled with 10 μl of propidium iodide (PI; 50 μg/ml; Sigma-Aldrich) at 37°C for 30 min and then analyzed using a BD FACSCalibur flow cytometer (BD Technologies, Carlsbad, CA, USA). The cell proportions in the G1, S, and G2/M phases were calculated using ModFit software (Verity Software House Inc., Topsham, ME, USA).
Nucleosomal Fragmentation Assay
Apoptosis was detected using a method described previously8. Molt 4 cells were treated with 10 μg/ml of vincristine (VCR) before transfection. At 48 h after transfection with RNA oligonucleotides, cell apoptosis was quantified by nucleosomal fragmentation (Cell Death Detection ELISA PLUS; Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s protocol. The absorbance values were normalized to those of control cells to derive a nucleosomal enrichment factor.
Migration and Invasion Assays
An 8-μm Transwell insert (Costar, Dallas, TX, USA) was used for the migration assay. A total of 5 × 103 cells was suspended in a serum-free medium and seeded into the upper chamber of the Transwell, and RPMI-1640 medium containing 100 ng/ml CXCL12 was added to the lower chamber. For invasion assays, 1 × 104 cells were suspended in serum-free medium and added to the upper chamber of 8-μm pore size Transwells precoated with 200 μl of Matrigel (BD Bioscience) at a concentration of 200 μg/ml. The medium with 100 ng/ml of CXCL12 was added to the lower chamber as a chemoattractant. After 24 h of incubation, the filters were fixed in methanol and stained with 4′-6-diamidino-2-phanylindole (DAPI; Sigma-Aldrich). After cells that failed to migrate or invade through the pores were carefully removed, five random fields were counted per chamber under an inverted florescent microscope (Carl-Zeiss, Berlin, Germany).
Western Blotting Assay
Proteins were extracted, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto nitrocellulose membranes (Millipore, Bedford, MA, USA). Western blotting was performed with primary antibodies against CXCR7, STAT3, phosphorylated (p)-STAT3 (Tyr-705), cyclin D1, Bcl-xl, Bcl-2 (Abcam, Cambridge, UK) and matrix metalloproteinase-2 (MMP-2), MMP-9, and β-actin (Abnova, Taiwan, P.R. China), followed by the secondary antibody conjugated with horseradish peroxidase (Sigma-Aldrich). Immunoreactivity was visualized by enhanced chemiluminescence kit (Santa Cruz Biotechnology, Santa Cruz, CA, USA) in accordance with the manufacturer’s instruction.
Terminal Transferase-Mediated dUTP Nick End Labeling (TUNEL) Assay
TUNEL assay was performed according to the manufacturer’s protocol. In brief, tissue specimens were fixed with 10% formalin overnight, embedded with paraffin, nonserially sectioned (4 μm), and mounted onto polylysine-coated slides. The sections were deparaffinized in xylene and rehydrated in a graded series of ethanol solutions. Afterward, they were rinsed with PBS and incubated with FITC-labeled terminal deoxynucleotidyl transferase nucleotide mix (Roche, Mannheim, BW, Germany) at 37°C for 60 min according to the manufacturer’s protocol. Subsequently, the sections were washed twice in PBS and counterstained with 10 mg/ml of DAPI. TUNEL+ cells were imaged and mounted using a fluorescence microscope (Carl-Zeiss). These cells were expressed as a percentage of the total cells determined by DAPI staining.
In Vivo Tumor Growth, Metastasis, and Apoptosis Assays
Animal experiments were officially approved by the Institutional Committee for Animal Research and performed in conformity with the national guidelines for the care and use of laboratory animals. For tumor growth assays, 6-week-old female mice with severe combined immune deficiency (SCID; Institute of Zoology, Chinese Academy of Sciences, Beijing, P.R. China) received subcutaneous injections of 5 × 106 Molt 4-luc cells with stably expressing miR-101 or shCXCR7 or their controls (n = 6 mice/group) through the hind flank. All mice were euthanized with sodium pentobarbital at 50 days postinoculation, and tumors were removed and weighed. One half of the tumor tissues were immediately stored at −80°C for qPCR and Western blot analyses. The residual tissues were immerged in paraffin, sectioned, and stained with TUNEL kits for apoptosis assay. TUNEL+ cells were examined and calculated under a fluorescence microscope (Carl-Zeiss). For tumor metastasis assays, mice were injected with 1 × 106 Molt 4-luc cells with a stable expression of miR-101 or shCXCR7 or their controls through the tail vein to establish the tumor metastasis model. Mice were sacrificed at 5 weeks postinjection, and lungs were removed and embedded in paraffin. Three nonsequential sections per animal were obtained. Sections were stained with hematoxylin/eosin (Maixin Biotech, Fuzhou, Fujian, P.R. China) and analyzed for metastasis by light microscopy (Carl-Zeiss). The total number of metastases per lung section was determined and averaged.
Bioluminescence Imaging and Quantification
Bioluminescence imaging was performed according to the manufacturer’s protocol. Six-week-old female SCID mice were subcutaneously injected with 5 × 106 Molt 4-luc cells infected with control lentivirus or lentivirus expressing miR-101 or shCXCR7. At 14 days after treatment, tumor growth was monitored through in vivo luciferase imaging of the xenografts. Mice were intraperitoneally injected with d-luciferin (Promega) at 150 mg/kg per mouse and anesthetized during image acquisition using the Xenogen IVIS imaging system. Signals in defined regions of interest were quantified as luminescence radiance (photons/s/cm2/sr) using Living Image software (Xenogen Corporation, Berkeley, CA, USA).
Statistical Analysis
Data are expressed as means ± standard deviation (SD) from three independent experiments. Data were analyzed by Student’s t-test and analysis of variance (ANOVA). Pearson correlation analysis was conducted to assess the statistical significance between cells with high or low levels of miR-101 or CXCR7. All p values were two-sided and obtained using SPSS 13.0 software package (SPSS Inc., Chicago, IL, USA). A value of p < 0.05 was considered statistically significant.
RESULTS
Inverse Correlation Between CXCR7 and miR-101 in T-ALL Samples
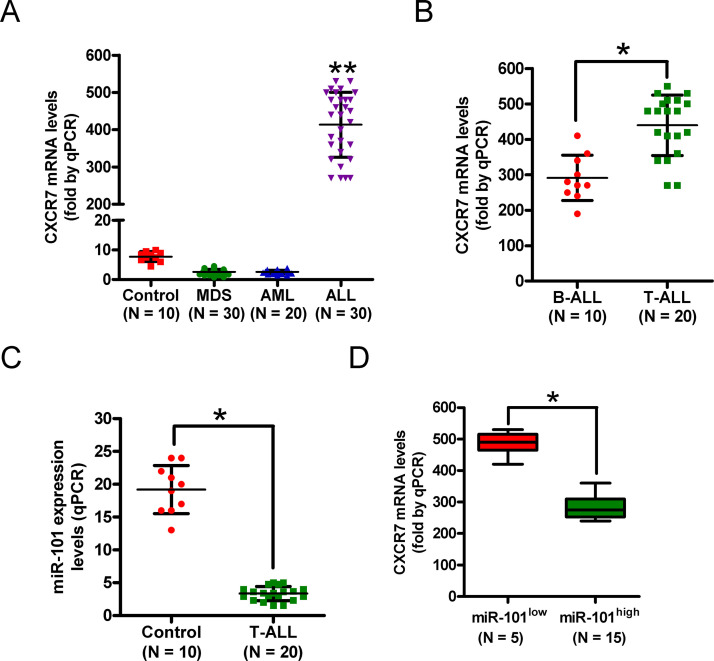
Clinically, we found that CXCR7 mRNA was highly upregulated in BM samples of ALL patients compared with those from healthy donors and MDS and AML patients (Fig. 1A). Among ALL-diagnosed patients, CXCR7 mRNA levels in the T-ALL subtype were higher than those in the B-ALL subtype (Fig. 1B). By contrast, qPCR analysis showed that miR-101 expression was lower in T-ALL samples than in specimens from healthy donors (Fig. 1C). Intriguingly, CXCR7 level inversely correlated with miR-101 level in T-ALL patients (Fig. 1D). These findings suggest that miR-101 expression is negatively associated with CXCR7 level in T-ALL specimens, and miR-101 and CXCR7 are possibly key players in T-ALL.
Figure 1.
The levels of C-X-C chemokine receptor type 7 (CXCR7) mRNA and microRNA-101 (miR-101) in T-cell acute lymphoblastic leukemia (T-ALL) samples. (A) Quantitative real-time polymerase chain reaction (qPCR) analysis of CXCR7 mRNA levels in bone marrow (BM) samples from ALL patients (n = 30) compared with those from healthy donors (control) and myelodysplastic syndrome (MDS; n = 30) and acute myeloid leukemia (AML) patients (n = 20). (B) mRNA levels of CXCR7 in B-ALL (n = 10) and T-ALL (n = 20) specimens were measured by qPCR assays. GAPDH was used as the endogenous control. (C) qPCR analysis of miR-101 levels in T-ALL samples (n = 20) compared with those from healthy donors (control; n = 10). U6 was used as the internal control. (D) Correlation between CXCR7 level and miR-101 expression in T-ALL specimens. All data are shown as mean ± SD of three separate experiments. *p < 0.05 versus B-ALL group or control group or miR-101low group; **p < 0.01 versus control group or MDS group or AML group.
Reduced miR-101 Expression and Increased CXCR7 Levels in T-ALL Cells
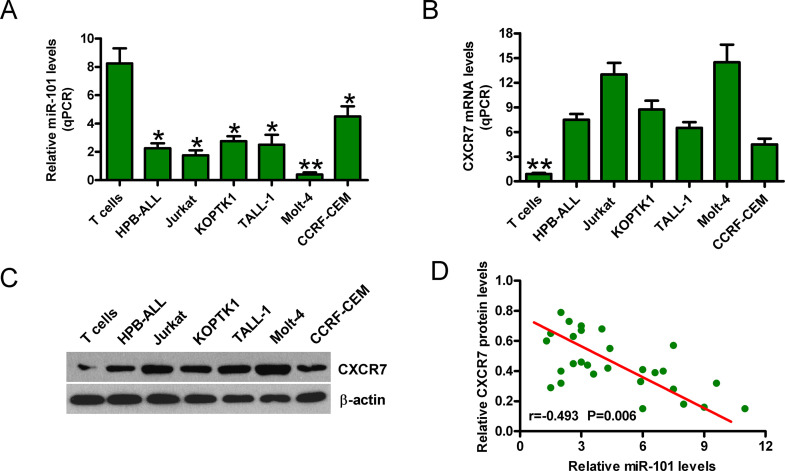
Next, we found that miR-101 levels were significantly lower in T-ALL cells (HPB-ALL, TALL-1, KOPTK1, Jurkat, CCRF-CEM, and Molt 4) than that in normal T cells (Fig. 2A). qPCR and Western blot analyses showed that CXCR7 expression was enhanced in T-ALL cell lines compared with normal T cells (Fig. 2B and C). Moreover, CXCR7 expression was inversely correlated with miR-101 level in T-ALL cell lines (Fig. 2D). The Molt 4 cell line with the lowest level of miR-101 and the highest level of CXCR7 was selected for further studies. These results indicate that miR-101 and CXCR7 play key roles in T-ALL development.
Figure 2.
The expression of miR-101 and CXCR7 in T-ALL cell lines. qPCR assays were conducted to assess miR-101 expression (A) and CXCR7 mRNA levels (B) in T-ALL cell lines (HPB-ALL, TALL-1, KOPTK1, Jurkat, CCRF-CEM, and Molt 4) and normal T cells. U6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as the internal controls, respectively. (C) Protein levels of CXCR7 in T-ALL cell lines and normal T cells were determined by Western blot. β-Actin was used as the endogenous control. (D) Inverse correlation between miR-101 and CXCR7 levels in T-ALL cell lines. All data are shown as mean ± SD of three separate experiments. *p < 0.05, **p < 0.01 versus normal T-cell group or all T-ALL cell groups.
miR-101 Directly Targets CXCR7 in T-ALL Cells
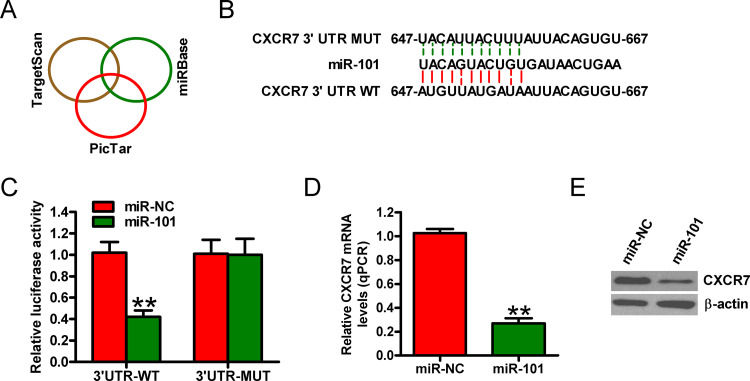
The targets of miR-101 were predicted using three algorithms: TargetScan, PicTar, and miRBase (Fig. 3A). Figure 3B shows a complementary sequence of miR-101 to the 3′-UTR of CXCR7 mRNA. Dual-luciferase reporter assay revealed that miR-101 decreased the activity of the luciferase reporter fused to the 3′-UTR-WT of CXCR7 but did not inhibit that of the reporter fused to the MUT version (Fig. 3C). Moreover, the introduction of miR-101 reduced the mRNA and protein levels of CXCR7 in Molt 4 cells (Fig. 3D and E). These data demonstrate that miR-101 directly targets CXCR7 in T-ALL cells.
Figure 3.
Identification of CXCR7 as a direct target of miR-101. (A) Putative target genes of miR-101 were predicted using TargetScan, PicTar, and miRBase software. (B) Predicted binding sites of miR-101 in the mutant (MUT) and wild-type (WT) 3′-UTR of CXCR7. (C) Dual-luciferase reporter assays were performed 24 h after cotransfection of Molt 4 cells with miR-NC or miR-101 mimics and a pGL3 construct containing the WT or MUT 3′-UTR of CXCR7. Data were normalized to those from cells cotransfected with miR-NC and pGL3 plasmid. mRNA (D) and protein (E) levels of CXCR7 in Molt 4 cells transfected with miR-NC or miR-101 mimics were detected by qPCR and Western blot assays. GAPDH and β-actin were used as internal controls. All data are shown as mean ± SD of three separate experiments. **p < 0.01 versus miR-NC group.
miR-101 Inhibits T-ALL Cell Proliferation, Apoptosis Resistance, Migration, and Invasion by Targeting CXCR7
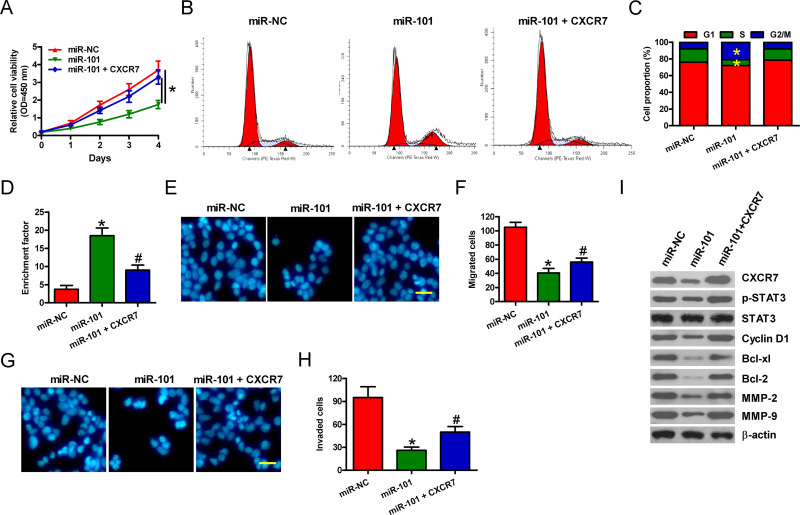
We investigated whether CXCR7 is a functional target of miR-101. Molt 4 cells were treated with miR-101 mimic or miR-NC or miR-101 mimic + CXCR7-expressing plasmid in the presence of 100 ng/ml of CXCL12. Cell viability significantly decreased in the miR-101-transfected cells compared with that in the miR-NC-treated cells, which was markedly attenuated by CXCR7 overexpression (Fig. 4A). In Molt 4 cells, the increase in G2/M phase and decrease in S phases by miR-101 were counteracted by CXCR7 restoration (Fig. 4B and C). As shown in Figure 4D, miR-101 augmented the percentage of apoptotic Molt 4 cells pretreated with 10 μg/ml of VCR, which was neutralized by CXCR7 overexpression. Whether miR-101 suppresses the migration and invasion of T-ALL cells by targeting CXCR7 was evaluated by Transwell assays. As expected, miR-101 inhibited the migration of Molt 4 cells, whereas CXCR7 overexpression partially counteracted the decrease (Fig. 4E and F). Similarly, the invasion of Molt 4 cells was reduced by miR-101, and the inhibitory effect was attenuated by CXCR7 restoration (Fig. 4G and H). Next, we explored the molecular mechanism by which miR-101 exerts its tumor-suppressive effects. The activation of STAT3, which is the predominant mediator of CXCR7 signaling, was reduced by miR-101 treatment, whereas CXCR7 overexpression attenuated the decrease in p-STAT3 expression (Fig. 4I). Moreover, the protein levels of STAT3 targets, including cyclin D1, Bcl-xl, Bcl-2, MMP-2, and MMP-9, were also detected. We found that miR-101 repressed the expressions of proteins mentioned above, which were counteracted by CXCR7 restoration (Fig. 4I). These results illuminate that miR-101 suppresses the proliferation, apoptosis resistance, migration, and invasion of T-ALL cells by blocking CXCR7/STAT3 signaling.
Figure 4.
miR-101 reduces proliferation, apoptosis resistance, and motility of T-ALL cells by targeting CXCR7. Molt 4 cells were transfected with miR-NC, miR-101 mimics, or miR-101 mimics + CXCR7-expressing plasmid. (A) CCK-8 assay was carried out to determine cell viability. (B, C) Flow cytometry analysis of Molt 4 cell fraction in G1, S, and G2/M phases 48 h after transfection. (D) Molt 4 cells subjected to VCR pretreatment were transfected with miR-NC or miR-101 mimics or miR-101 mimics + CXCR7-expressing plasmid. Apoptosis was measured by nucleosomal fragmentation assay. Migration assay was performed for Molt 4 cells. Images were captured after 24 h of transfection (E), and the number of migrated cells was calculated (F). Cell invasion was detected using Transwell chambers (G), and the number of invaded cells was quantified (H). (I) Representative Western blot results of p-signal transducer and activator of transcription 3 (STAT3), STAT3, Cyclin D1, Bcl-xl, Bcl-2, matrix metalloproteinase (MMP-2), and MMP-9. β-Actin was used as a loading control. All data are shown as mean ± SD of three replicates. *p < 0.05 versus miR-NC group; #p < 0.05 versus miR-101 group.
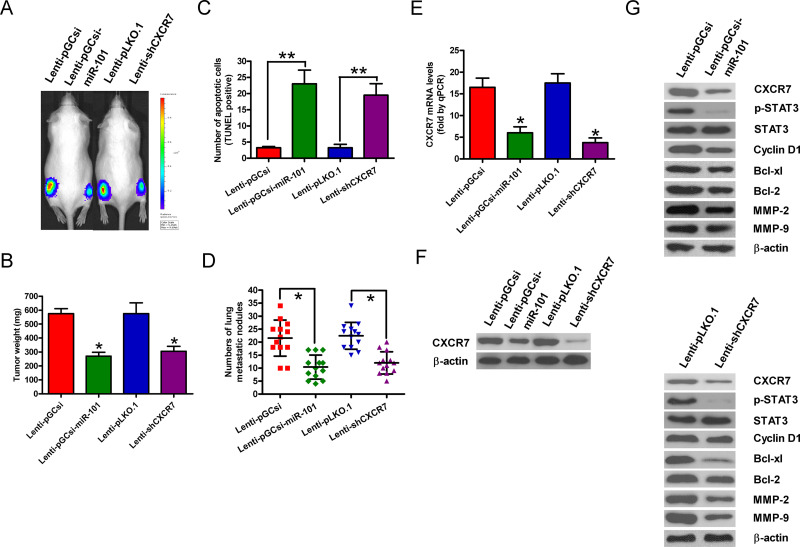
miR-101 Overexpression or CXCR7 Depletion Retards Tumorigenesis and Metastasis of T-ALL In Vivo
A xenograft or metastasis mouse model was established by the subcutaneous or intravenous injection of Molt 4-luc cells stably expressing an miR-101 precursor or shCXCR7 or their vector controls. Compared with the control groups, the miR-101 overexpression or CXCR7 knockdown group exhibited significant reductions in tumor growth (Fig. 5A) and weight (Fig. 5B). Apoptotic cells were much more in the tumors of the ectopic miR-101 expression or CXCR7-silencing group than those in the tumors derived from the control groups (Fig. 5C). miR-101 restoration or CXCR7 depletion also effectively retarded lung metastases in vivo (Fig. 5D). In addition, the mRNA and protein levels of CXCR7 were significantly reduced in miR-101-overexpressed or CXCR7-silenced tumor tissues (Fig. 5E and F). Mechanistically, the levels of p-STAT3, cyclin D1, Bcl-xl, Bcl-2, MMP-2, and MMP-9 were decreased in the tumors of the miR-101-overexpressed or CXCR7-depleted group (Fig. 5G). These results indicate that the suppression of T-ALL growth and lung metastasis by miR-101 is mediated by CXCR7 reduction in vivo.
Figure 5.
miR-101 overexpression or CXCR7 knockdown suppressed tumor growth and metastasis in vivo. Severe combined immune deficiency (SCID) mice were injected subcutaneously or intravenously with Molt 4-luc cells that were infected with a control lentivirus (Lenti-pGCsi or Lenti-pLKO.1) or a recombinant lentivirus expressing an miR-101 precursor (Lenti-pGCsi-miR-101) or shCXCR7 (Lenti-shCXCR7). (A) In vivo luciferase image for the detection of xenograft growth. (B) Tumor weight was measured after 50 days of implantation. (C) Terminal transferase-mediated dUTP nick end labeling (TUNEL) assay was conducted to detect the percentage of apoptotic cells. (D) Numbers of metastatic foci in the lungs from various groups at 5 weeks after tail vein injection. mRNA (E) and protein (F) levels of CXCR7 in the tumor tissues were assessed by qPCR and Western blot assays. GAPDH and β-actin were used as the loading controls, respectively. (G) Representative Western blot results of p-STAT3, STAT3, cyclin D1, Bcl-xl, Bcl-2, MMP-2, and MMP-9 in the tumor tissues. β-Actin was used as a loading control. All data are shown as mean ± SD of three separate experiments. *p < 0.05, **p < 0.01 versus Lenti-pGCsi or Lenti-pLKO.1 group.
DISCUSSION
miRNA and gene expression profiles have provided valuable insights into the molecular mechanisms of leukemia20. In this study, we found that miR-101 is downregulated in T-ALL cells and samples, negatively associating with CXCR7 expression. Integrative bioinformatics prediction and dual-luciferase reporter assay indicated that CXCR7 is a direct target of miR-101. Functionally, the inhibitory effects of miR-101 on T-ALL cell proliferation, apoptosis resistance, and motility were reversed by CXCR7 restoration in vitro. miR-101 overexpression and CXCR7 depletion significantly reduced the malignant characteristics of T-ALL in vivo. Overall, these results suggest that miR-101 acts as a tumor suppressor in T-ALL by targeting CXCR7.
Reportedly, miR-101 is silenced in numerous solid tumors6–8. However, the expression of miR-101 in T-ALL has rarely been identified. Herein, we demonstrated that miR-101 was lowly expressed in T-ALL cell lines and patient samples. Li et al.8 revealed that miR-101 elicits oncostatic effects on breast cancer. Consistently, we showed that miR-101 significantly reduced T-ALL cell proliferation, migration, and invasion. Qian et al.12 illuminated that miR-101 regulates T-ALL progression and chemotherapeutic sensitivity. Interestingly, we also found that miR-101 abolished apoptosis resistance of T-ALL cells to VCR. These findings indicate that miR-101 acts as a suppressor in T-ALL carcinogenesis.
Many studies have revealed that miR-101 targets different effectors, including CXCR76–8. As an important chemokine receptor, CXCR7 elevation frequently emerges and promotes the growth and metastasis of malignancies13–19. The upregulation of CXCR7 cell surface expression was also highlighted in T-ALL cells18. We demonstrated that, in opposition to miR-101, CXCR7 was significantly upregulated in T-ALL specimens. Similarly, our investigation on the intrinsic function of CXCR7 in T-ALL revealed that CXCR7 counteracted the inhibitory effects of miR-101 on the proliferation, apoptosis resistance, and motility of human T-ALL cells in vitro, which is a direct supportive evidence for CXCR7 function. Li et al.8 reported that CXCR7 is highly important for breast cancer growth and metastasis in vivo, and their findings are supported by human breast cancer xenograft and metastatic models. Similarly, we found that CXCR7 depletion markedly reduced the malignant characteristics of T-ALL in vivo. These results indicated that CXCR7 was a functional mediator of miR-101 in T-ALL.
Recently, Li et al.8 demonstrated that CXCR7 promotes proliferation and motility of breast cancer cells via activation of STAT3 signaling. Actually, STAT3 is often overexpressed in many types of leukemia, including T-ALL21. A previous study employing STAT3-deficient mice showed that STAT3 activation is responsible for bypassing apoptosis and enhancing T-cell proliferation, suggesting that inhibition of STAT3 activation can be an effective therapeutic approach for T-ALL22. STAT3 activation is associated with proliferation, apoptosis resistance, cell cycle progression, and invasion of T-ALL cells23–25. Constitutively active STAT3 induces the transcription of a number of target genes, including cyclin D1, Bcl-xl, Mcl-1, MMP-2, and MMP-924–26. STAT3 has been shown to promote cell cycle progression and apoptosis resistance by increasing cyclin D1, Bcl-xl, and Mcl-1 expression in T-ALL cells25. MMP-2 and MMP-9 are the matrix metalloproteinases that break down the extracellular matrix27 and are implicated in cell migration and invasion in acute leukemia28. Consistently, in this study, we demonstrated that miR-101 reduced phosphorylation of STAT3, accompanied with reduction of cyclin D1, Bcl-xl, Bcl-2, MMP-2, and MMP-9. Similar to the effects of miR-101 overexpression, CXCR7 depletion abolished the expression of p-STAT3 and STAT3 target genes. These results indicate that miR-101 hinders the CXCR7/STAT3 signaling pathway in T-ALL.
Our study has limitations in deciphering the role of the CXCL12/CXCR7 axis in miR-101-downregulated T-ALL. In this study, we only illuminated the promoting effects of CXCR7/STAT3 signaling on T-ALL malignancies but did not identify whether other downstream molecules of CXCR7 are involved in the progression of T-ALL and their biological functions. Moreover, we did not investigate whether miR-101 alone or CXCR7 alone or the combination of miR-101 and CXCR7 can be a potential prognostic indicator in T-ALL patients. It needs more studies to verify them.
In summary, miR-101 is downregulated in T-ALL. In vitro and in vivo studies confirmed that miR-101 is a novel suppressor of T-ALL by directly targeting CXCR7. Overall, these findings suggest that miR-101 impedes T-ALL growth and metastasis by targeting CXCR7 and highlight the potential role of miR-101/CXCR7 in therapeutic use of T-ALL patients.
ACKNOWLEDGMENTS
We appreciated that the Blood Department of the 150th Central Hospital of PLA provided the samples.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350(15):1535–48. [DOI] [PubMed] [Google Scholar]
- 2. Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, Cohen HJ, Sallan SE, Asselin BL. Childhood T-cell acute lymphoblastic leukemia: The Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21(19):3616–22. [DOI] [PubMed] [Google Scholar]
- 3. Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, Biondi A, Pui CH, Downing JR, Campana D. Early T-cell precursor leukaemia: A subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009;136(2):215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene 2006;25(46):6170–5. [DOI] [PubMed] [Google Scholar]
- 6. Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, Ye ZY, Tao HQ. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer 2010;46(12):2295–303. [DOI] [PubMed] [Google Scholar]
- 7. Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69(3):1135–42. [DOI] [PubMed] [Google Scholar]
- 8. Li JT, Jia LT, Liu NN, Zhu XS, Liu QQ, Wang XL, Yu F, Liu YL, Yang AG, Gao CF. MiRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7. Oncotarget 2015;6(31):30818–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robertus JL, Kluiver J, Weggemans C, Harms G, Reijmers RM, Swart Y, Kok K, Rosati S, Schuuring E, van Imhoff G, Pals ST, Kluin P, van den Berg A. MiRNA profiling in B non-Hodgkin lymphoma: A MYC-related miRNA profile characterizes Burkitt lymphoma. Br J Haematol. 2010;149(6):896–9. [DOI] [PubMed] [Google Scholar]
- 10. Papakonstantinou N, Ntoufa S, Chartomatsidou E, Papadopoulos G, Hatzigeorgiou A, Anagnostopoulos A, Chlichlia K, Ghia P, Muzio M, Belessi C, Stamatopoulos K. Differential microRNA profiles and their functional implications in different immunogenetic subsets of chronic lymphocytic leukemia. Mol Med. 2013;19:115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Correia NC, Melao A, Povoa V, Sarmento L, Gomez de Cedron M, Malumbres M, Enguita FJ, Barata JT. microRNAs regulate TAL1 expression in T-cell acute lymphoblastic leukemia. Oncotarget 2016;7(7):8268–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qian L, Zhang W, Lei B, He A, Ye L, Li X, Dong X. MicroRNA-101 regulates T-cell acute lymphoblastic leukemia progression and chemotherapeutic sensitivity by targeting Notch1. Oncol Rep. 2016;36(5):2511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hattermann K, Held-Feindt J, Lucius R, Muerkoster SS, Penfold ME, Schall TJ, Mentlein R. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 2010;70(8):3299–308. [DOI] [PubMed] [Google Scholar]
- 14. Iwakiri S, Mino N, Takahashi T, Sonobe M, Nagai S, Okubo K, Wada H, Date H, Miyahara R. Higher expression of chemokine receptor CXCR7 is linked to early and metastatic recurrence in pathological stage I nonsmall cell lung cancer. Cancer 2009;115(11):2580–93. [DOI] [PubMed] [Google Scholar]
- 15. Wang J, Shiozawa Y, Wang J, Wang Y, Jung Y, Pienta KJ, Mehra R, Loberg R, Taichman RS. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283(7):4283–94. [DOI] [PubMed] [Google Scholar]
- 16. Guo JC, Li J, Zhou L, Yang JY, Zhang ZG, Liang ZY, Zhou WX, You L, Zhang TP, Zhao YP. CXCL12-CXCR7 axis contributes to the invasive phenotype of pancreatic cancer. Oncotarget 2016;7(38):62006–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi A, Shi H, Dong L, Xu S, Jia M, Guo X, Wang T. CXCR7 as a chemokine receptor for SDF-1 promotes gastric cancer progression via MAPK pathways. Scand J Gastroenterol. 2017;52(6–7):745–53. [DOI] [PubMed] [Google Scholar]
- 18. Melo RCC, Longhini AL, Bigarella CL, Baratti MO, Traina F, Favaro P, de Melo Campos P, Saad ST. CXCR7 is highly expressed in acute lymphoblastic leukemia and potentiates CXCR4 response to CXCL12. PLoS One 2014;9(1):e85926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li W, Ding Q, Ding Y, Lu L, Wang X, Zhang Y, Zhang X, Guo Q, Zhao L. Oroxylin A reverses the drug resistance of chronic myelogenous leukemia cells to imatinib through CXCL12/CXCR7 axis in bone marrow microenvironment. Mol Carcinog. 2017;56(3):863–76. [DOI] [PubMed] [Google Scholar]
- 20. Zhao H, Wang D, Du W, Gu D, Yang R. MicroRNA and leukemia: Tiny molecule, great function. Crit Rev Oncol Hematol. 2010;74(3):149–55. [DOI] [PubMed] [Google Scholar]
- 21. Ferrajoli A, Faderl S, Ravandi F, Estrov Z. The JAK-STAT pathway: A therapeutic target in hematological malignancies. Curr Cancer Drug Targets 2006;6(8):671–9. [DOI] [PubMed] [Google Scholar]
- 22. Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: Generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161(9):4652–60. [PubMed] [Google Scholar]
- 23. Takemoto S, Mulloy JC, Cereseto A, Migone TS, Patel BK, Matsuoka M, Yamaguchi K, Takatsuki K, Kamihira S, White JD, Leonard WJ, Waldmann T, Franchini G. Proliferation of adult T cell leukemia/lymphoma cells is associated with the constitutive activation of JAK/STAT proteins. Proc Natl Acad Sci USA 1997;94(25):13897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishikawa C, Arbiser JL, Mori N. Honokiol induces cell cycle arrest and apoptosis via inhibition of survival signals in adult T-cell leukemia. Biochim Biophys Acta 2012;1820(7):879–87. [DOI] [PubMed] [Google Scholar]
- 25. Xiao R, Gan M, Jiang T. Wogonoside exerts growth-suppressive effects against T acute lymphoblastic leukemia through the STAT3 pathway. Hum Exp Toxicol. 2017;36(11):1169–76. [DOI] [PubMed] [Google Scholar]
- 26. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer 2009;9(11):798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roomi MW, Monterrey JC, Kalinovsky T, Rath M, Niedzwiecki A. Patterns of MMP-2 and MMP-9 expression in human cancer cell lines. Oncol Rep. 2009;21(5):1323–1333. [DOI] [PubMed] [Google Scholar]
- 28. Klein G, Vellenga E, Fraaije MW, Kamps WA, de Bont ES. The possible role of matrix metalloproteinase (MMP)-2 and MMP-9 in cancer, e.g. acute leukemia. Crit Rev Oncol Hematol. 2004;50(2):87–100. [DOI] [PubMed] [Google Scholar]