Abstract
Coronavirus disease- 2019 (COVID-19) has rapidly become a major threat to humans due to its high infection rate and deaths caused worldwide. This disease is caused by an RNA virus, Severe Acquired Respiratory Syndrome –Corona Virus-2 (SARS-CoV-2). This class of viruses have a high rate of mutation than DNA viruses that enables them to adapt and also evade host immune system. Here, we compared the first known Nucleocapsid Phosphoprotein (N protein) sequence of SARS-CoV-2 from China with the sequences from Indian COVID-19 patients to understand, if this virus is also mutating, as it is spreading to new locations. Our data revealed twenty mutations present among Indian isolates. Out of these, mutation at six positions led to changes in the secondary structure of N protein. Further, we also show that these mutations are primarily destabilising the protein structure. The candidate mutations identified in this study may help to speed up the understanding of variations occurring in SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, Mutations, Nucleocapsid Phosphoprotein (N), Infectious diseases, India
COVID-19; SARS-CoV-2; Mutations; Nucleocapsid Phosphoprotein (N); Infectious diseases; India
1. Introduction
In the Wuhan City of China (Hubei province), multiple cases of severe pneumonia like symptoms were reported for the first time in December 2019 [1]. Initially, it was thought to be a viral disease and after the complete genome sequencing, it was revealed that it belongs to a novel coronavirus and later renamed as SARS-CoV-2 because of its similarity with SARS-CoV [2]. The disease caused by SARS-CoV-2 has been termed Coronavirus disease-2019 (COVID-19). The COVID-19 has been declared by the World Health Organization (WHO) as a global pandemic in March 2020. As of 6th June 2020, there were more than 7 million confirmed cases of COVID-19 with 0.4 million confirmed deaths worldwide.
The SARS-CoV-2 is rapidly spreading to new locations across the globe and infecting human populations. The viruses have the tendency to mutate which helps them to survive better in the host. Most of the RNA viruses including coronaviruses have been reported to possess an inherent high rate of mutation [3]. Reports suggest that the Spike protein and RNA dependent RNA polymerase (RdRp) are two mutational hotspot proteins of SARS-CoV-2 that is frequently mutated as revealed by the sequencing data from different countries [4, 5, 6, 7].
One of the important proteins that protect RNA genome of SARS-CoV-2 is N protein [8]. The N protein comprises of three distinct conserved regions; the N-terminal domain (NTD), C-terminal domain (CTD) and intrinsically disordered regions (IDRs). There are three IDRs, namely at N-terminal end, C-terminal end and between the NTD and CTD [8]. All of these regions make contact with the viral genomic RNA. Further, evidences suggest that the NTD have various residues that are critical for its interactions with the RNA genome of SARS-CoV-2. The CTD is also known as dimerization domain because two molecules of N protein dimerise with the interactions of their CTD. The NTD and CTD are separated by serine and arginine rich, IDR also known as the linker region (LKR). The main function of N protein is to protect viral RNA genome of SARS-CoV-2 by proper packaging. It binds with the viral genomic RNA and generates long, relatively flexible and somewhat helical ribonucleoprotein complex also known as viral capsid [9].
In this study, we analysed the N protein sequences of SARS-CoV-2 from Indian COVID-19 patients and compared with the Wuhan virus sequence. Our study identified twenty mutations in N protein among Indian isolates, and discussed their possible consequences.
2. Material and methods
2.1. N protein sequence retrieval
As of 7th June 2020, there were 175 protein sequences of SARS-CoV-2 deposited in NCBI- virus database from India. We downloaded these sequences and their protein identification accession number are mentioned in supplementary table 1. The N protein sequence from the earliest sequenced virus from Wuhan wet seafood market area was used as a reference (accession number: YP_009724397).
2.2. Multiple sequence alignments
We used Clustal Omega online tool to perform multiple sequence alignment (MSA) to identify mutations present among Indian isolates as described earlier [10]. The Indian sequences were compared with the Wuhan N protein (accession number: YP_009724397). The mutant residues were carefully identified and marked. Clustal Omega tool offers fast and reliable algorithm to perform sequence alignments to efficiently compare thousands of input sequences [11].
2.3. Secondary structure prediction
For secondary structure prediction, we used Chou and Fasman secondary structure prediction (CFSSP) online tool [12]. This tool provides the secondary structure pattern from the input primary amino acid sequence.
2.4. Calculations of difference in free energy (ΔΔG) between wild-type and mutant
The ΔΔG was calculated using online mCSM webserver [13]. It uses an algorithm to calculate the differences in free energy between the wild-type and mutant protein. The positive ΔΔG indicates stabilising mutation while negative ΔΔG represents destabilisation. mCSM tool also shows the impact of a mutation on the atomic-distance patterns surrounding an amino acid residue. For this analysis RCSB protein ID: 6VYO [14] was used for NTD molecular modelling and RCSB protein ID: 6WJI (Centre for Structural Genomics of Infectious Diseases-CSGID, unpublished) was used for CTD molecular modelling of N protein.
2.5. Analysis of protein dynamicity
We analysed the impact of mutation on dynamicity and structural variations using DynaMut webserver [15]. This webserver provides the location of a selected amino acid in the 3D-structure of the protein and also show the kind of interactions made by the residues with surrounding amino acids. For this analysis, the crystal structure of N protein was first uploaded on DynaMut webserver. The RCSB protein ID: 6VYO [14] and 6WJI were used for NTD and CTD molecular modelling of N protein respectively. It also provides the differences in the vibrational entropy between wild-type and mutant protein and predicts the impact of mutation on structural stability and dynamicity.
3. Results
3.1. N protein of SARS-CoV-2 is rapidly mutating in India
We downloaded all N protein sequences from India deposited in NCBI virus database till 7th June 2020. The accession numbers of the N protein used in this study are mentioned in supplementary table 1. The multiple sequence alignments were performed using Clustal Omega tool and the Wuhan virus N protein sequences was used as a reference (accession number: YP_009724397). Our data revealed twenty mutations present in the SARS-CoV-2 N protein from Indian COVID-19 patients (Figure 1). The complete details of mutations identified in this study are listed in supplementary table 2. Further, these mutations are spreading all over the N protein; however, a cluster is observed in the intrinsically disordered region 2 (IDR2) or linker region (LKR) present between NTD and CTD (Figure 1). There are seven mutations present at this location suggesting that this might be a mutational hotspot area of this protein. The N-terminal unstructured region harbours three mutations at 6, 13 and 33 positions. The NTD and CTD have various mutations as shown in Figure 1. Subsequently, we also analysed the frequency of twenty mutations among Indian isolates (Table 1). Our data revealed S194L and P13L are most frequently mutated while rest of the mutants have low frequency (Table 1). Altogether, we have identified twenty mutations in N protein present among Indian isolates.
Figure 1.
Mutational analysis of SARS-CoV-2 sequences reported from India. The sequence of SARS-CoV-2 N protein obtained from Indian COVID-19 patients was aligned with the sequence from Wuhan (wet seafood market) SARS-CoV-2. The mutations were recognised by amino acid sequence alignment by Clustal Omega. i) The mutant residues positions and mutations are highlighted in the schematic of N protein. The N protein sequence of Wuhan SARS-CoV-2 was used as reference for this analysis. ii) The schematic of N protein showing different domains. N-terminal domain (NTD), C-terminal domain (CTD), intrinsically disordered regions 1, 2, 3 (IDR1, IDR2 and IDR3), IRD2 also known as linker region (LKR) because it connects NTD and CTD. NTD have RNA binding motif while CTD helps in protein dimerization.
Table 1.
Mutations across N proteins of SARS-CoV-2 among Indian isolates and their frequencies. The frequency of each mutation was calculated by counting the number of samples that exhibited the variation among 175 samples analysed in this study. Similarly, the percentage frequency was also calculated.
| Mutation | Frequency | % Frequency |
|---|---|---|
| P6T | 2 | 1.142857 |
| P13L | 16 | 9.142857 |
| S33I | 2 | 1.142857 |
| R92S | 1 | 0.571429 |
| G120R | 1 | 0.571429 |
| L139F | 1 | 0.571429 |
| A152S | 1 | 0.571429 |
| A156S | 1 | 0.571429 |
| R191L | 1 | 0.571429 |
| S194L | 49 | 28.0 |
| S202N | 5 | 2.857143 |
| R203K | 5 | 2.857143 |
| G204R | 5 | 2.857143 |
| M234I | 1 | 0.571429 |
| G236C | 1 | 0.571429 |
| P302S | 1 | 0.571429 |
| P344S | 1 | 0.571429 |
| D348Y | 1 | 0.571429 |
| T362I | 1 | 0.571429 |
| T393I | 1 | 0.571429 |
3.2. Mutations residing in NTD and CTD of N protein are leading to change in the secondary structure
Subsequently, to understand the possible implication of the identified mutations on N protein structure, we evaluated the secondary structure of mutant residues and compared with the wild-type sequence. Since, NTD and CTD only have defined secondary structure; therefore, we analysed only those mutations which reside in these two domains of N protein. Out of the 20 mutations identified in this study, nine mutations are present in NTD and CTD. Our data revealed that mutation at six positions alter the secondary structure as shown in Figure 2. We have used CFSSP webserver to computationally analyse the probable effect of mutation on secondary structure. Each panel of Figure 2 comprises of two parts (i and ii), part i represents Wuhan isolate (wild type sequence) and part ii represents Indian Isolate (mutant sequence). Both part i and ii shows the contribution of individual amino acid to the secondary structure of N protein (H- helix, T-turn, E-beta sheet). The location of mutant residue is highlighted in red box for better comparisons. Overall, Figure 2 suggests the effect of identified mutation on the secondary structure of the N protein where the mutant residue resides. Our data suggest that mutation at 92, 152 and 156 (Figure 2A, D and E; compare panel i with ii) positions in NTD led to alteration in the secondary structure. Similarly, mutation at position 344, 348 and 362 (Figure 2G, H and I; compare panel i with ii) alters the secondary structure in CTD. Among these, there is a major shift in the secondary structure at 92 position which harbours arginine to serine mutation. Due to this mutation the beta-sheet structure is completely replaced by coiled-coil secondary structure (Figure 2A, compare panel i with ii). Altogether, our data strongly suggest that the mutations identified in this study led to alteration in secondary structure that might affect the functioning of N protein.
Figure 2.
Prediction of the secondary structure of N protein. The panels A, B, C, D, E, F, G, H and I indicate nine mutations observed among Indian isolates that are localised in NTD and CTD of N protein. Panel A (R92S), B (G120R), C (L139F), D (A152S), E (A156S) and F (P302S) represent NTD mutations while, G (P344S), H (D348Y) and I (T362I) represent mutations in the CTD of N protein. In each panel, (i) represents the sequence of Wuhan isolate and (ii) represents sequence of Indian isolates. Both part (i) and (ii) shows the contribution of individual amino acid to the secondary structure of N protein (H- helix, T-turn, E-beta sheet). The location of mutant residue is highlighted in red box for better comparisons. We observed change in the secondary structure between Wuhan, and Indian isolate at six mutation sites (Panel A, D, E, G, H and I). The figure of each panel (A–I) was obtained from CFSSP webserver.
3.3. Mutant N proteins have altered protein stability and dynamicity
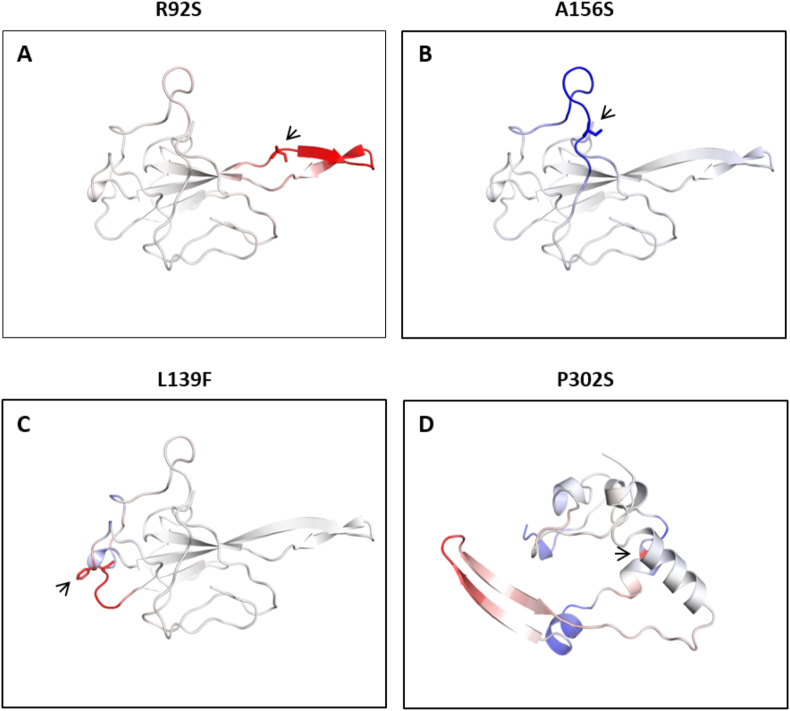
Since the secondary structures were changed due to the mutations; therefore, we decided to study the effect of these mutations on the stability of the N protein. We used mCSM tool that provide the impact of mutation on the dynamic structure by calculating the difference in free energy (ΔΔG) between wild-type and mutant protein. The values more than zero indicates stabilising mutation and values below zero represents destabilising mutation. The analyses were done with those mutations that are present in the NTD and CTD of N protein. As shown in Table 2, the mutations are leading to the destabilisation of protein structure, and the effect was considerably higher at four positions namely, R92S, L139F, A156S and P302S. The ΔΔG for these four positions are negative in the range of 1.0–2.0 kcal.mol−1 (Table 2). Subsequently, we used DynaMut webserver to perform protein modelling to analyse the overall protein structural flexibility upon mutation at four positions that exhibited the maximum change in ΔΔG. Our data revealed that R92S led to gain in flexibility (Figure 3A) and A156S increased the rigidification (Figure 3B). However, L39F and P320S mutations caused both rigidity and flexibility in some areas of the protein structure (Figure 3C and D).
Table 2.
Calculations of ΔΔG between wild-type and mutant Nucleocapsid Phosphoprotein. mCSM webserver was used to calculate the predicted ΔΔG. The negative values indicate the destabilization of protein upon mutation.
| S. No. | PDB File | Wild-type Residue | Residue Position | Mutant Residue | Predicted ΔΔG (kcal.mol−1) | Outcome |
|---|---|---|---|---|---|---|
| 1 | 6vyo.pdb | R | 92 | S | -1.146 | Destabilizing |
| 2 | 6vyo.pdb | G | 120 | R | -0.432 | Destabilizing |
| 3 | 6vyo.pdb | L | 139 | F | -0.94 | Destabilizing |
| 4 | 6vyo.pdb | A | 152 | S | -0.3 | Destabilizing |
| 5 | 6vyo.pdb | A | 156 | S | -1.422 | Destabilizing |
| 6 | 6wji.pdb | P | 302 | S | -1.475 | Destabilizing |
| 7 | 6wji.pdb | P | 344 | S | -0.346 | Destabilizing |
| 8 | 6wji.pdb | D | 348 | Y | -0.006 | Destabilizing |
| 9 | 6wji.pdb | T | 362 | I | -0.1 | Destabilizing |
Figure 3.
Prediction of the impact of mutations on structural integrity of N protein. Panel A, B, C and D represent the site of respective mutations. The panel A (R92S), B (A156S) and C (L139F) shows the mutations that are localised in the NTD while panel D (P302S) represents CTD mutant. In each panel, the blue color signifies a rigidification in the protein structure, and red represents the gain in flexibility upon mutation. The molecular structures of N protein shown in each panel were downloaded from DynaMut webserver. The arrowhead indicates the location of mutant amino acid in the protein structure.
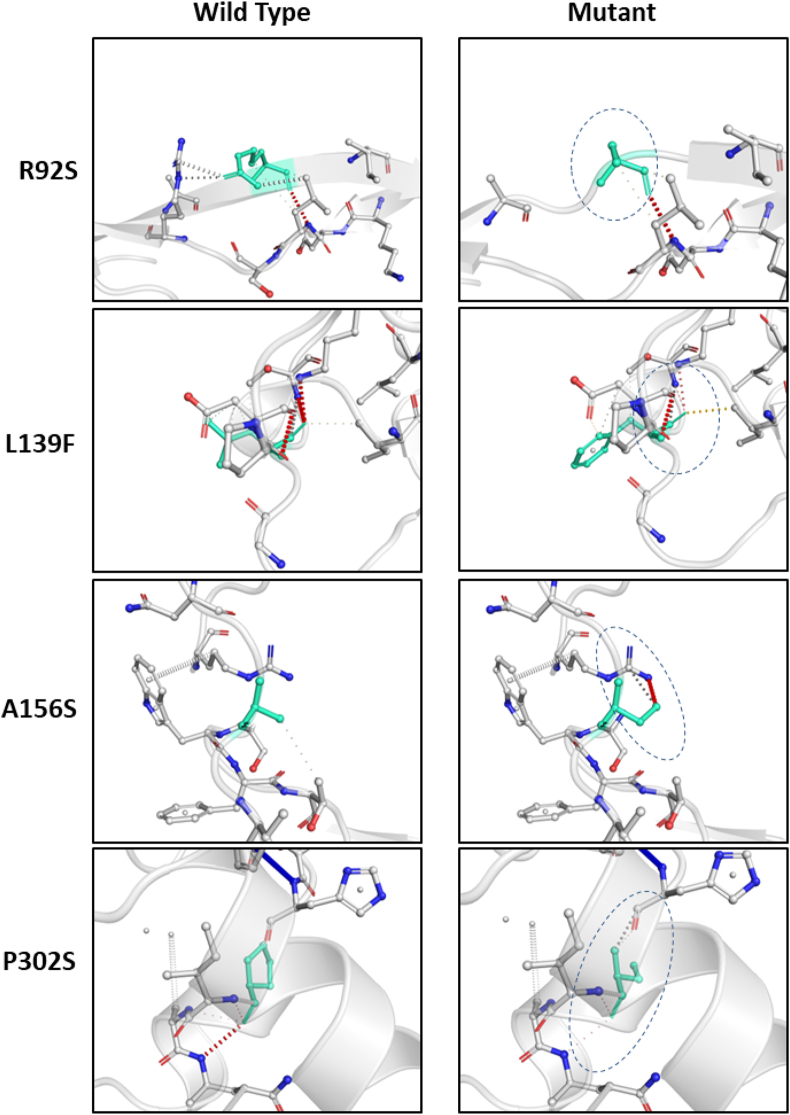
Since, our data show that mutations are altering ΔΔG as well as the protein dynamicity (rigidity and flexibility). To substantiate these data, we sought to analyse the impact of the mutations on intramolecular interactions made by mutant amino acids in the N protein. Our analysis revealed that the mutations were notably affecting the intramolecular bonds in the pockets where these amino acids reside (Figure 4). Specifically, the R92S mutant lost almost all interactions with its neighbouring amino acids (Figure 4). Minor changes in intramolecular interactions were also visible at other mutant positions (Figure 4). Altogether, our data strongly suggest that N protein mutations led to alteration in its structure and dynamicity.
Figure 4.
Visual representation of close range intramolecular interactions contributed by the highlighted residues in their respective three-dimensional positions. The molecular structures of wild-type and mutant amino acids of N protein are highlighted in the light-green color. The surrounding residues which are making close contacts with the wild-type and mutant residues are also depicted. The localisation of four mutants is shown from top to bottom that includes R92S, A156S, L139F and P302S respectively. Panel in the left side represents wild-type while the right side represents mutant version. The structures were downloaded from DynaMut webserver. The dashed circle highlights the variation in intramolecular bonds between the wild-type and mutant structure.
4. Discussions
The SARS-CoV-2 is an RNA virus and its genome is replicated by RdRp that have weak proofreading activity. Therefore, the RNA viruses have relatively higher level of mutations than DNA viruses as they replicate [3]. These mutations enable them to survive in the host cells and also adapt them to new locations and environmental conditions. The SARS-CoV-2 pandemic rapidly spread after the first reported case from Wuhan, China and within 3 months it reached to almost all countries worldwide. As the virus migrated to new locations they also attained mutations. In order to understand the mutations occurring in India, we compared the N protein sequences of SARS-CoV-2 from Indian patients with the earliest known sequence from China. Our data revealed twenty mutations among Indian isolates that are distributed all over the protein. However, we found a distinct cluster in the LKR which is an IDR between the NTD and CTD (Figure 1). IDRs do not have a well-defined tertiary structure in the native form; however, IDRs of coronavirus N proteins play important role in binding with the viral genomic RNA [16]. LKR also plays primary role in the intracellular signalling [17, 18, 19]. Further, we report multiple mutations that reside in the NTD which are known to make contact with the RNA genome of this virus. Previous study shows that the Arg-76 in the infectious bronchitis virus (IBV) N protein is a well conserved across the coronavirus family and required for efficient RNA binding, and may structurally resemble the Arg-94 in the SARS-CoV N protein [20], and possibly Arg-92 of SARS-CoV-2 N protein. Our study revealed that Arg-92 is mutated to Serine and therefore, it might affect N protein and RNA interactions.
We also observed mutations in CTD that could possibly affect its dimerization potential. Earlier report shows that 302, 347, 352, 358 and other residues are involved in dimerization in SARS-CoV N protein [21]. This study identified mutations at 302, 344, 348 and 362 residues which are in the close vicinity of these critical residues. Overall, our data predicts the effect of mutation on the stability of the N protein. Although, we lack ‘experimental validation’ of our computational data; however, based on the computational methods, we have identified twenty mutations and concluded that among those twenty, four mutations (R92S, L139F, A156S and P302S) are most likely to affect the structure and stability of the N protein. The secondary structure prediction tool shows that mutation at several locations (92, 152 and 156) will either favour helix/beta sheet or turn structure. This means that these mutations might cause shift in the secondary structure that can affect the stability of the mutant protein. Subsequently, the stability of protein was computationally analysed by DynaMut protein stability prediction tool.
Reports suggest that coronavirus N proteins also acts as an RNA chaperones [22, 23] and helps in the maintenance and proper folding of the RNA genome. Therefore, drugs that can interfere with N protein function are of great pharmacological interests. The drugs that inhibit oligomerisation of N protein via its CTD are promising candidates to inhibit virus assembly [24]. A recent study with SARS-CoV-2 N protein identified ten promising drugs that can inhibit its function including Conivaptan, Ergotamine, Venetoclax, Rifapentine and others [25]. Alanine at 156th position is among the critical residues that interacts with these promising drug candidates [25]. Our study shows mutation at 156th position; thereby, it could possibly cause alteration in drug binding pocket of N protein. Therefore, we predict that few of these mutants might have differential interaction with the drugs, and that can possibly contribute to the drug resistance.
Declarations
Author contribution statement
Gajendra Kumar Azad: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to acknowledge Patna University, Patna for infrastructural support.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018 doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laamarti M., Jr., Medical R. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geodistribution and a rich genetic variations of hotspots mutations. Biorxiv. 2020 doi: 10.1371/journal.pone.0240345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020 doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chand G.B., Banerjee A., Azad G.K. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ. 2020;8 doi: 10.7717/peerj.9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chand G.B., Banerjee A., Azad G.K. Identification of twenty-five mutations in surface glycoprotein (Spike) of SARS-CoV-2 among Indian isolates and their impact on protein dynamics. Gene Rep. 2020;21:100891. doi: 10.1016/j.genrep.2020.100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein - Forms and functions. Antivir. Res. 2014 doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surjit M., Lal S.K. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect. Genet. Evol. 2008 doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azad G.K. Identification of novel mutations in the methyltransferase complex (Nsp10-Nsp16) of SARS-CoV-2. Biochem. Biophys. Rep. 2020 doi: 10.1016/j.bbrep.2020.100833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashok Kumar T. Wide Spectr; 2013. CFSSP: Chou and Fasman Secondary Structure Prediction Server. [Google Scholar]
- 13.Pires D.E.V., Ascher D.B., Blundell T.L. MCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang C.-K., Hsu Y.-L., Chang Y.-H., Chao F.-A., Wu M.-C., Huang Y.-S., Hu C.-K., Huang T.-H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory Syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009 doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stohlman S.A., Baric R.S., Nelson G.N., Soe L.H., Welter L.M., Deans R.J. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J. Virol. 1988 doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker M.M., Masters P.S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990 doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014 doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C.Y., ke Chang C., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., huang Huang T. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007 doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zúñiga S., Cruz J.L.G., Sola I., Mateos-Gómez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010 doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zúñiga S., Sola I., Moreno J.L., Sabella P., Plana-Durán J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007 doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo Y.S., Lin S.Y., Wang S.M., Wang C.T., Chiu Y.L., Huang T.H., Hou M.H. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013 doi: 10.1016/j.febslet.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadioglu O., Saeed M., Greten H.J., Efferth T. Identification of novel compounds against three targets of SARS CoV2 coronavirus by combined virtual screening and supervised machine learning. Bull. World Health Organ. 2020 doi: 10.1016/j.compbiomed.2021.104359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.