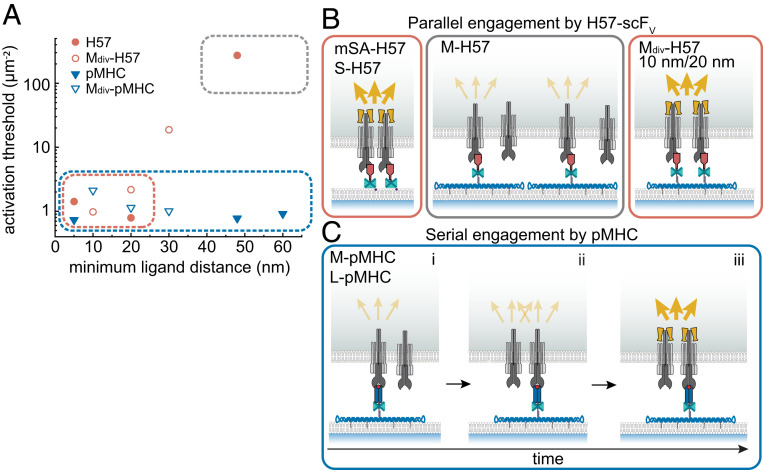
Fig. 5.
Ligand-specific spatial requirements for T cell activation. (A) Minimum ligand distances δ are plotted versus the activation threshold. For DNA origami platforms featuring a single ligand, δ corresponds to the smallest possible distances between two ligands on adjacent platforms assuming quasi-crystalline packing in clusters. For divalent platforms, it corresponds to the predefined distance between the two ligands on an individual platform. The lateral extension of mSA-anchored ligands was approximated with 5 nm. The spatial regimes allowing (red box) and prohibiting (gray box) parallel engagement via H57 as well as those that allow serial engagement via pMHC (blue box) are indicated. (B) Parallel engagement model of antibody-induced triggering. Two or more triggered TCRs within 20 nm form signaling-competent TCR assemblies, resulting in ZAP-70 recruitment (yellow dumbbells) and initiation of calcium signaling (yellow arrows). H57-scFV–triggered T cell activation requires at least two simultaneous ligand–receptor engagements within a distance of 20 nm (“parallel engagement”). This can occur via T cell–induced clustering of mSA-anchored scFVs or preorganization of scFVs on divalent DNA origami platforms (red boxes). H57-scFvs isolated on M-DNA origami platforms at δ = 48 nm fail to stimulate T cells efficiently (gray box). (C) Serial engagement model of pMHC-induced triggering (blue box). A single isolated pMHC molecule can create a signaling-competent TCR assembly by sequentially engaging multiple TCRs. (i) A single pMHC–TCR binding event does not induce efficient downstream signaling. (ii) Via serial, short-lived binding events, a single isolated pMHC molecule may engage several TCRs sequentially. (iii) If two or more TCRs within a distance of 20 nm are triggered in this way, a signaling-competent TCR assembly is created.