Fig. 1.
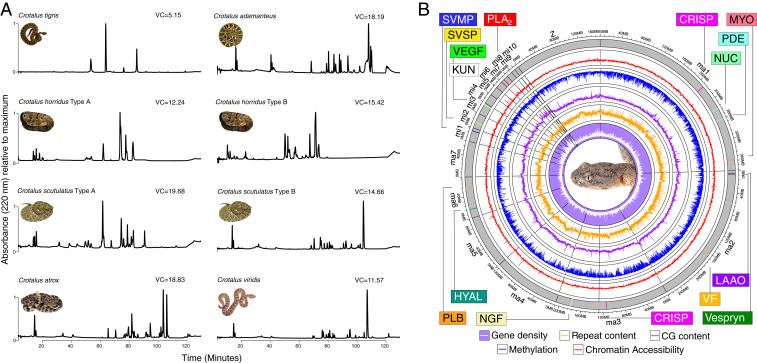
A complex genetic architecture underlies the simple venom phenotype of the Tiger Rattlesnake. (A) Comparative proteomics shows the simplicity of the Tiger Rattlesnake (C. tigris) venom phenotype. RP-HPLC of whole venoms for eight individuals across six species of rattlesnake. Chromatographic peaks represent a particular toxic protein or set of proteins. Estimates of VC, our proxy for phenotypic complexity, are shown next to each venom profile, and the Tiger Rattlesnake (VC = 5.15) possessed a significantly simpler venom than all other rattlesnakes shown here (see SI Appendix, Table S1 for details). For species polymorphic for the A–B venom dichotomy, where type A venoms are simple and neurotoxic and type B venoms are complex and hemotoxic, venom type is listed next to species name. C. tigris possesses a type A venom, whereas Crotalus adamanteus, C. atrox, and C. viridis possess type B venoms. See SI Appendix, Fig. S4 for transcriptome–proteome relationship in C. tigris. (B) Circos plot of the RaGOO assembly displaying gene density, repeat content, GC content, proportion of methylated cytosines in the venom gland, and chromatin accessibility in the venom gland across 100-kb windows at chromosome scale. Venom toxins that were reliably mapped to chromosome scaffolds are shown as colored vertical lines and labeled accordingly. CRISP, cysteine-rich secretory protein; HYAL, hyaluronidase; KUN, Kunitz-type toxin; LAAO, L-amino acid oxidase; MYO, myotoxin; NGF, nerve growth factor; NUC, nucleotidase; PDE, phosphodiesterase; PLB, phospholipase B; VEGF, vascular endothelial growth factor; VF, venom factor. Snake image credits: M.P.H. and Travis Fisher (photographer).