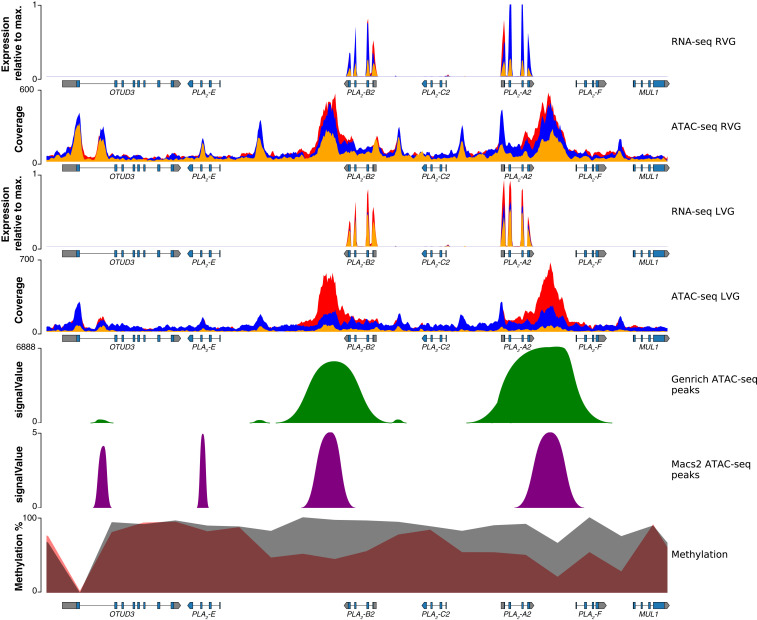
Fig. 4.
Regulatory landscape for the toxin-gene family in the Tiger Rattlesnake. Chromatin accessibility and methylation levels regulate the expression of toxin genes with increased accessibility and reduced methylation near active genes (i.e., -B2 and -A2) and reduced accessibility and increased methylation near silenced genes. Rows from top to bottom: right venom-gland (RVG) transcriptome, RVG ATAC-seq, left venom-gland (LVG) transcriptome, LVG ATAC-seq, venom-gland specific ATAC-seq peaks identified by Genrich, venom-gland specific ATAC-seq peaks identified by MACS2, and methylation percentage. RNA-seq y axes represent venom-gland expression levels scaled by the most highly expressed transcript in that genomic region. Red, blue, and orange colors in the RNA-seq and ATAC-seq plots represent the three individuals sequenced, respectively; colors are overlaid, and not all may be visible. Genrich-estimated signalValues represent the area under the peak; MACS2-estimated signalValues represent fold change in coverage at the peak summit. For the methylation track, gray shading represents methylation levels in 2-kb bins in the pancreas (control), whereas red shading represents methylation levels in 2-kb bins in the venom gland. Gene annotations are shown under tracks where appropriate; -B2, -C2, and -A2 are toxin genes. Max., maximum.