Abstract
Cancer stem cells (CSCs) are a small cell subpopulation in many cancer types and are involved in various processes of tumor progression, such as initiation, metastasis and recurrence. The distinguished features of CSCs include a variety of biological properties, including self-renewal, multidifferentiation, stemness marker expression, and resistance to chemotherapy and radiotherapy. Despite their great potential of clinical importance, the CSC signaling pathways are not well understood at the molecular level. MicroRNAs (miRNAs) are a class of endogenous noncoding RNAs that play an important role in the regulation of several cellular, physiological, and developmental processes. Aberrant miRNA expression is associated with many human diseases, including cancer. miRNAs have been implicated in the regulation of CSC properties; therefore, a better understanding of miRNA-induced modulation of CSC gene expression could aid in the identification of promising biomarkers and therapeutic targets. In the present review, we summarize the major findings of the impacts of miRNAs on CSC signaling networks; we then discuss the recent advances that have improved our understanding of CSC regulation by miRNA-mediated signaling networks and that may lead to the development of miRNA therapeutics specifically targeting CSCs.
Keywords: microRNA, Cancer stem cells (CSCs), Tumor initiation, Therapeutic resistance, Metastasis and recurrence
1. Introduction
The cancer stem cell (CSC) theory was initially proposed more than 150 years ago [1,2], and the basis of this theory is that cancer cells are derived from stem cell populations in tissues. Although there are conflicting data about cancer stem cells, tumor-initiating cells, and the origin of cancer cells, accumulating evidence suggests that so-called CSCs and normal stem cells have many similarities with regard to biological properties, e.g., self-renewal, differentiation capacity and specific marker expression [3]. The differences between CSCs and normal tissue stem cells are tumorigenicity and the chemoresistance ability. Although some exceptions have been reported [4], the CSC theory is generally accepted for various types of cancers in both the basic research and cancer therapeutic fields.
Common signaling pathways and networks were found in CSCs and normal stem cells. These pathways and networks, including Wnt/β-catenin, JAK/STAT, PI3/AKT, and NF-kB, are known to regulate stem cell self-renewal and differentiation. Abnormalities in the Wnt/β-catenin pathway were shown to enhance self-renewal in leukemia stem cells [5]. In the case of myxoid liposarcoma, the JAK-STAT pathway regulates cancer stem cell properties such as chemoresistance [6].
MicroRNAs (miRNAs) are noncoding RNAs that have versatile functions in physiology and pathophysiology. It is well known that miRNAs play a pivotal role in cancer initiation and progression. On the other hand, several miRNAs are also known to regulate signaling pathways [7]. In this review, we discuss the recent findings of miRNA-related signaling pathways in cancer biology, particularly focusing on those involved in CSCs. A better understanding of the characteristics of CSCs in the biological properties of miRNA-related signaling pathways is important for basic science as well as clinical applications.
1.1. Biogenesis and functions of miRNAs
miRNAs are noncoding RNAs of 21–24 nucleotides that generally negatively regulate gene expression at the posttranscriptional level. Mechanistically, miRNAs directly bind to partially complementary sequences in the 3′-untranslated regions (3′UTRs) of target genes and lead to the degradation of target mRNAs or repression of mRNA translation [8]. The majority of miRNA biogenesis occurs by transcribing miRNAs by RNA polymerase II as pri-miRNAs, which are long primary transcripts, and miRNAs are processed in the nucleus by the RNase III Drosha in combination with cofactors such as DGCR8 into precursor miRNAs (pre-miRNAs) 70–100 nucleotides long. DGCR8 is an evolutionarily conserved protein that interacts with proline-rich peptides through its WW domain, and heterozygous deletion results in the most common human genetic deletion syndrome, known as DiGeorge syndrome [9]. The pre-miRNA is exported from the nucleus to the cytoplasm by exportin-5, a family member of RanGTP-binding transport receptors [10], and is finally cleaved in a complex composed of the RNase III Dicer into mature miRNAs. The mature miRNA (also called the guide strand of mature miRNA) is incorporated into an RNA-induced silencing complex (RISC). The RISC consists of Argonaute (Ago) proteins and GW182. Although miRNAs basically act as negative regulators of target mRNAs by binding to the 3′UTRs of target mRNAs, it has also been reported that miRNAs can also bind to the 5′UTR or the open reading frame and regulate the translation of target mRNAs [11,12].
1.2. Relationship between cancer stem cells and miRNAs
Similar to protein-coding genes, abnormal expression of miRNAs has been reported in various types of cancers [13]. The functional investigation of miRNAs has clearly demonstrated that oncogenic miRNAs and tumor suppressor miRNAs play pivotal roles in cancer initiation and progression. Additionally, some miRNAs are known to be related to the regulation of CSC properties, such as asymmetric cell division, tumorigenicity, and drug resistance.
One of the most famous oncogenic miRNAs is the miR-17–92 cluster. The miR-17-92 cluster is polycistronically expressed from the chromosome 13q31 locus, which is known to be amplified in lung cancer [14]. The expression levels of miR-17–92 are higher in tumor tissues than in normal tissues, and increased miR-17–92 expression significantly promotes tumorigenesis in lymphoma [15]. Based on this evidence, it is considered an oncogenic miRNA. A bioinformatics analysis identified the tumor suppressor PTEN (phosphatase and tensin homolog deleted on chromosome ten) as a direct target gene of the miR-17–92 cluster [16]. Because PTEN induces apoptotic cell death via the P13K-Akt-PKB pathway, miR-17–92 indirectly regulates this signaling pathway.
Conversely, the expression levels of some miRNAs are decreased in tumorigenesis. Generally, this kind of miRNA is considered to work as a tumor suppressor miRNA. Tumor suppressor miRNAs negatively regulate oncogenes or oncogenic pathways and inhibit tumor development. The most classic and famous tumor suppressor miRNAs are the let-7 family [17]. Let-7 was originally discovered in Caenorhabditis elegans, and the let-7 sequences are conserved among species. It has been reported that let-7c suppresses the self-renewal of stem cells and inhibits the estrogen-induced activation of Wnt signaling in breast cancer stem cells [18]. Another well-known tumor suppressor miRNA, miR-34, which is a direct target of p53, was shown to inhibit Notch signaling pathways [19]. As such, in this review, we focus on the roles of miRNAs in the signaling pathways involved in CSC properties and summarize the therapeutic potential of miRNAs targeting CSCs.
2. Involvement of miRNAs in maintaining CSCs
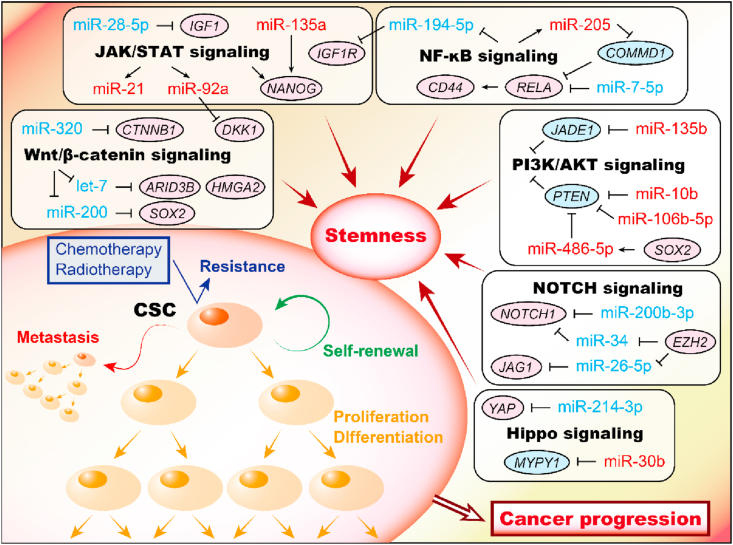
CSCs, which are characterized by the capacity for self-renewal and differentiation, were first identified in acute myeloid leukemia in 1997 [20,21]. CSCs usually share specific stemness-related markers, such as CD44 and CD133, but they can be different depending on the cancer type [[22], [23], [24], [25]]. Moreover, several stemness-related markers, such as NANOG, OCT4, SOX2, and KLF4, can be targeted by a single miRNA, namely, miR-145, in embryonic stem cells and CSCs [26,27]. Furthermore, various other miRNAs are involved in stemness by regulating several signaling pathways, including the Wnt/β-catenin, PI3K/Akt, and NF-kB pathways, and the detailed functions of the miRNAs in each signaling pathway are described below (Fig. 1).
Fig. 1.
Overview of miRNAs in CSCs. miRNAs which regulate signaling pathways and miRNAs which are regulated by CSC-related signal pathways are shown. As CSC-related pathways based on the literatures, Wnt/β-catenin signaling, JAK/STAT signaling, NF-κB signaling, PI3K/AKT signaling, NOTCH signaling and Hippo signaling were illustrated to interacted with miRNAs.
2.1. Wnt/β-catenin signaling
Wnt signaling is an evolutionarily conserved pathway that plays various important roles, including serving as a regulator of stem cell proliferation and self-renewal [28]. Wnt signaling through its β-catenin receptors is called the canonical signaling pathway, and the noncanonical signaling pathways are also functional [28]. Inhibition of Wnt/β-catenin signaling leads to the upregulation of pri-let-7a and pri-miR-200c [29]. The expression of let-7 is high in differentiated cancers, and let-7 decreases the expression of OCT4 and SOX2 by targeting the 3′-UTR of ARID3B and HMGA2 [30,31]. Conversely, LIN28B suppresses the expression of let-7 and induces stem-like genes [32]. Thus, the expression of the let-7 family is associated with favorable prognosis in patients with lung cancer [33]. Moreover, the miR-200 family can directly target SOX2, and in contrast, lncRNA Sox2ot promotes stemness by sponging the miR-200 family [34]. Furthermore, miR-200c suppresses clonogenicity by targeting BMI1 [35]. Therefore, let-7 and the miR-200 family suppress stemness properties.
On the other hand, circ_001680 exerts its oncogenic function, including stemness, by sponging miR-340, which also targets BMI1 [36]. Moreover, miR-22 represses the expression of miR-200 and induces stemness by directly targeting the TET family [37]. Conversely, lncRNA MIR22HG suppresses Wnt/β-catenin signaling by modulating both mature miR-22-3p and miR-22-5p [38]. In addition, miR-600, miR-128-3p, miR-302c, and miR-320 also weaken stemness by targeting the mediators of Wnt/β-catenin signaling SCD1, NEK2, CARF, and CTNNB1, respectively [[39], [40], [41], [42]]. Hypoxia is also a key promoter of CSCs, and miR-1275 is regulated by HIF-1α and maintains stemness by targeting multiple genes in the Wnt signaling pathway, such as DKK3, SFRP1, GSK3β, and RUNX3 [43]. Therefore, various miRNAs and competing endogenous RNAs (ceRNAs) are involved in the regulation of Wnt/β-catenin signaling.
2.2. JAK/STAT signaling
JAK/STAT signaling has crucial roles in inflammation, proliferation, and survival. Several cytokines and growth factors, such as IL-6, IGF, and TGF-β1, can activate the pathway through their receptors, and nuclear STAT mediates transcriptional regulation of various target genes [44].
First, regarding stemness, nuclear CD44/acetylated STAT3 signaling is required for tumorigenicity and can be associated with metastasis [45]. In addition, IGF-1 is one of the key stimulators of signaling, and downregulation of miR-28-5p promotes CSC self-renewal by targeting IGF-1 [46]. As a downstream target of IGF/STAT3 signaling, NANOG is activated, and moreover, miR-135a enhances the expression of NANOG by modulating methylation of the promoter region [47,48]. Then, STAT3 signaling further increases the expression of miR-21 and miR-92a, and they induce stemness by modulating Wnt signaling [[49], [50], [51]]. However, IL-6/STAT3 signaling suppresses miR-34a, resulting in cancer progression [52].
Furthermore, radiation promotes the release of extracellular vesicles containing miR-603, and miR-603 then confers stemness properties and radioresistance by targeting both IGF and IGF1R [53]. On the other hand, the miR-1181 directory suppresses SOX2 and STAT3, and miR-7 suppresses STAT3 by targeting SETDB1 [54,55].
Therefore, JAK/STAT3 signaling is also important for stemness and is regulated by several oncogenic and tumor suppressor miRNAs. Moreover, STAT3 is also involved in the regulation of NF-κB signaling, which is discussed in the next section.
2.3. NF-κB signaling
The NF-κB family consists of the following five transcribed genes: p65 (RelA), RelB, c-Rel, p105/p50, and p100/p52, which are critical regulators of inflammation and immune responses [56]. In addition, NF-κB is activated downstream of oncogenic pathways, such as the RAS, BCR-ABL, p53, and PTEN pathways [56].
In stemness-enriched cancer cells, miR-205 upregulated by NF-κB signaling suppresses COMMD1, and then, downregulation of COMMD1 promotes intrinsic and TNF-α-induced inflammatory responses and RelA expression to maintain signaling activation [57]. RelA promotes CD44 expression but is targeted by the tumor suppressor miR-7-5p [58]. Previously, we identified that CD44 induces miR-629-3p expression and confers cisplatin resistance [59]. Moreover, NF-κB suppresses the expression of miR-194-5p by directly binding to its promoter region, and downregulation of miR-194-5p contributes to tumor progression by targeting IGF1R and PPFIBP1 [60]. Therefore, NF-κB is also important for cancer development, including stemness.
2.4. PI3K/AKT signaling
PI3K/AKT signaling regulates key metabolic processes, including glucose, lipid, protein, and nucleotide synthesis [61]. In addition to the direct metabolic functions of AKT, it also activates key downstream effectors, such as the mTORC1, GSK3 and FOXO transcription factors [61].
The AKT/mTOR pathway is activated by miR-135b and increases the expression of stemness markers [62]. However, PI3K signaling is attenuated by PTEN, and thus, miR-10b promotes CSC self-renewal by targeting PTEN [61,63]. Moreover, miR-106b-5p increases the expression of p-Akt by targeting PTEN and p21, resulting in resistance to irradiation [64]. Furthermore, SOX2 directly binds to the promoter region of miR-486-5p, and increased expression of miR-486-5p also suppresses expression of PTEN and FOXO1 [65]. Therefore, these miRNAs promote stemness by targeting negative regulators of PI3K signaling.
In addition, PD-L1 expression is positively correlated with stemness marker expression, and miR-873 inactivates PI3K/Akt and ERK1/2 signaling by directly targeting PD-L1 [66]. Moreover, miR-1976 suppresses CSC properties by targeting PIK3CG, and higher expression of miR-1976 is associated with favorable prognosis in patients with triple-negative breast cancer [67].
Therefore, aberrant PI3K/AKT signaling, which is frequently observed in various cancers, is also important for the maintenance of stemness.
2.5. Notch signaling
Notch signaling, which usually requires cell–cell contact, is essential for crosstalk between cancer cells and other components of the tumor microenvironment [68]. Notch signaling is one of the regulators of the CSC division mode, symmetrically or asymmetrically, and symmetric division is associated with high self-renewal ability and exponential tumor growth [69,70]. Mechanistically, miR-200b-3p targets NOTCH1 and inhibits symmetric division of CSCs by modulating the Notch/Numb ratio [69]. Moreover, lncRNA TUSC-7 increases the expression of NUMB by sponging miR-146 and inactivates Notch signaling [70]. Therefore, NOTCH1 promotes symmetric division by producing two daughter cells with high NOTCH1 expression, while Numb promotes asymmetric division.
Notch signaling also promotes multidrug resistance in various cancers, and miR-34, which is suppressed by EZH2-mediated H3 lysine 27 trimethylation (H3K27me3), targets Notch1 in CSCs [71,72]. Moreover, the tumor suppressors miR-21-5p and miR-26a-5p are also silenced by H3K27me3, and thus, EZH2 can be one of the targets to inactivate NOTCH signaling and weaken self-renewal [71,73]. Similarly, the expression of miR-7-5p is regulated by DNA methylation in the promoter region, and its downregulation promotes tumor progression by targeting Hes1, a key effector of Notch signaling [74]. Moreover, NOTCH2/3 is targeted by miR-195-5p, miR-181b, and miR-136 [[75], [76], [77]].
In addition, according to a double-negative feedback loop of the miR-200 family and ZEB1 in epithelial–mesenchymal transition, ZEB1 can indirectly activate Notch signaling because the miR-200 family has several other target genes involved in Notch signaling, such as Jag1, Maml2, and Maml3 [78]. Interestingly, exosomes can also activate Notch signaling. Exosomes containing miR-142-3p are secreted by bone marrow-derived mesenchymal stem/stromal cells (BM-MSCs) in the tumor microenvironment, and miR-142-3p promotes Notch signaling by downregulating Numb in CSCs [79]. Therefore, Notch signaling does not necessarily require cell–cell contact and is deeply involved in stemness.
2.6. Hippo signaling
Hippo signaling is involved in organ development, tissue regeneration, and tumorigenesis, including stemness [80]. YAP and TAZ are two main downstream targets of signaling, and YAP is highly expressed in the self-renewal of embryonic stem cells [80].
In lung squamous cell carcinoma, high expression of miR-214-3p is associated with favorable prognosis and suppresses tumorigenesis by targeting YAP1 [81]. On the other hand, miR-30b enhances CSC-like properties by targeting MYPT1, an upstream gene of the Hippo signaling pathway [82]. Importantly, YAP1 regulates miRNA biogenesis depending on the cell density, and at a low cell density, YAP1 activation results in global miRNA suppression [83]. Therefore, the Hippo pathway can be a strong regulator of the miRNA-regulated stemness described in this review. These miRNAs were listed in Table 1.
Table 1.
Stemness associated miRNAs and their target genes.
| miRNA | target gene | cell type | Reference |
|---|---|---|---|
| let-7 | ARID3B and HMGA2 | Oral SCC | [11] |
| let-7 family | HMGA2 | Ovarian cancer | [12] |
| miR-106b | PTEN and p21 | Colorectal cancer | [45] |
| miR-10b | PTEN | Breast cancer | [44] |
| miR-1181 | SOX2 and STAT3 | Pancreatic cancer | [36] |
| miR-1275 | DKK3, SFRP1, GSK3β, RUNX3, and NUMB | Lung adenocarcinoma | [24] |
| miR-128-3p | NEK2 | Breast cancer | [20] |
| miR-135a | DNMT1 | Liver cancer | [29] |
| miR-135b | JADE1 | Pancreatic cancer | [43] |
| miR-136 | NOTCH3 | Ovarian cancer | [57] |
| miR-142-3p | NUMB | Colon cancer | [60] |
| miR-145 | OCT4, SOX2, and KLF4 | Human embryonic stem cells | [7] |
| miR-145 | NANOG, OCT4, and SOX2 | Uterine cervical SCC | [8] |
| miR-146 | NUMB | Lung adenocarcinoma | [51] |
| miR-181b | NOTCH2 and RBPJ | NSCLC | [58] |
| miR-194-5p | IGF1R and PPFIBP | Ovarian cancer | [41] |
| miR-195-5p | NOTCH2 and RBPJ | Colorectal cancer | [56] |
| miR-1976 | PIK3CG | Breast cancer | [48] |
| miR-200 family | SOX2 | Pancreatic ductal adenocarcinoma | [15] |
| miR-200 family | JAG1, MAML2, and MAML3 | Several cell lines | [59] |
| miR-200b-3p | NOTCH1, TRIM2, PROX1, and NUMB | Pancreatic cancer | [50] |
| miR-200c | BMI1 | Breast cancer | [16] |
| miR-205 | COMMD1 | Head and Neck SCC | [38] |
| miR-21 | TGFβR2 | Colon cancer | [30] |
| miR-214-3p | YAP1 | Lung SCC | [62] |
| miR-21-5p | JAG1 | Hepatocellular carcinoma | [54] |
| miR-22 | TET family | Breast cancer | [18] |
| miR-22-3p | SFRP2 | Glioblastoma | [19] |
| miR-22-5p | PXDH15 | Glioblastoma | [19] |
| miR-26a-5p | JAG1 | Hepatocellular carcinoma | [54] |
| miR-28-5p | IGF-1 | Liver cancer | [27] |
| miR-302c | CARF | Colon cancer | [21] |
| miR-30b | MYPT1 | Ovarian cancer | [63] |
| miR-320 | CTNNB1 | Prostate cancer | [23] |
| miR-340 | BMI1 | Colorectal cancer | [17] |
| miR-34a | IL-6R | Colorectal cancer | [33] |
| miR-34a | NOTCH1, NOTCH2, and JAG1 | Cholangiocarcinoma | [52] |
| miR-34a | NOTCH1 | Breast cancer | [53] |
| miR-486-5p | PTEN and FOXO1 | Glioblastoma | [46] |
| miR-600 | SCD1 | Breast cancer | [22] |
| miR-603 | MGMT | Glioblastoma | [34] |
| miR-7 | SETDB1 | Breast cancer | [35] |
| miR-7 | RELA, Slug, and LncRNA XIST | Breast cancer | [39] |
| miR-7-5p | SMO and HES1 | Gastric cancer | [55] |
| miR-873 | PD-L1 | Breast cancer | [47] |
| miR-92a | DDK1 | Ovarian cancer | [32] |
SCC, squamous cell carcinoma; NSCLC, non-small cell lung cancer.
3. The therapeutic potential of miRNAs targeting CSCs
Because several miRNAs typically work as oncogenic or tumor suppressor miRNAs in various types of cancers, targeting miRNAs, i.e., blocking oncogenic miRNAs or enhancing tumor suppressor miRNAs in cancer, is considered to be a novel type of cancer therapy [84]. As described above in this review, some miRNAs that can regulate CSC properties could serve as therapeutics.
One of the challenges of realizing miRNA-based CSC therapeutics is the protection of miRNAs from RNase degradation in serum. To overcome this issue, there are two major strategies: one is to use chemically modified nucleic acids for higher stability, and the other is to establish an effective delivery system for miRNAs [85,86]. To increase the stability and overexpress tumor suppressor miRNAs in cancer cells, miRNA mimics are usually modified with methylation, such as 2′-O-methylation, of the passenger strand. In contrast, to block oncogenic miRNAs, inhibitors of miRNAs (also known as antimiRs) are often used by modification with locked nucleic acid (LNA) technology. Additionally, for miRNA therapeutics, tremendous efforts have been made to improve the delivery system of miRNAs using liposomes. Although the utility of liposomes in vivo has not been clinically feasible because of low uptake efficiency and high cytotoxicity, technological advancements in miRNA stability and delivery efficiency have been accomplished in the past several decades by merging biology and chemistry.
It has been reported that extracellular vesicles such as exosomes are considered to be suitable carriers for miRNA delivery in vivo. Exosomes are small extracellular vesicles with a diameter of approximately 50–150 nm. Exosomes are released into the extracellular microenvironment to transfer their components, including miRNAs, proteins and metabolites [87]. Thus, the application of exosomes as drug and gene therapy delivery systems is very promising as a natural vector system. Additionally, emerging evidence has demonstrated that surface proteins such as integrins define the organotropism of exosome delivery [88]; thus, modifying surface protein patterns on exosomes may improve targeted delivery systems of miRNAs. More recently, Usman et al. showed that antimiR against miR-125b was efficiently packaged with an electroporation method into exosomes collected from human red blood cells [89]. Local injection of antimiR in a breast cancer mouse model and systemic injection of antimiR in an AML mouse model confirmed the potential utility of antimiR-packaged exosomes for cancer therapeutics.
Considering CSC-targeted miRNA therapeutics, one of the promising targets is let-7 miRNA. The Let-7 family includes ten human isoforms, and they generally target a variety of oncogenes. Differentiated cancer cells express high levels of let-7, and loss of let-7 promotes tumor progression by suppressing RAS signaling [90]. Additionally, since let-7 targeted ARID3B and HMGA2 and induced CSC differentiation, enhanced expression of let-7 could suppress the CSC population in bulk tumor tissues [30].
4. Conclusion
Technical advances in single-cell approaches have clearly demonstrated the intratumoral heterogeneity of cancer cells and a unique subpopulation that is capable of tumor reconstitution. Several signaling pathways, such as the Wnt/β-catenin, PI3K/Akt, and NF-kB pathways, are well known to be involved in CSC maintenance and differentiation. A number of studies presented in this review have shown that miRNAs can work as regulators of these pathways and act as oncogenic or tumor suppressor miRNAs in CSCs. As such, miRNAs could be therapeutic targets for CSCs, e.g., inhibition of certain miRNAs might eradicate CSCs in tumor tissues. Collectively, a better understanding of miRNA functions associated with CSC properties could provide new insight into cancer therapeutics that possibly improve patient prognosis and treatment outcomes.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Project for Cancer Research and Therapeutic Evolution (P-CREATE; grant number:17cm0106402h0002) and Takeda Science Foundation.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
References
- 1.Cohnheim J. Ueber entzundung und eiterung. Path Anat Physiol Klin Med. 1867;40:1–79. [Google Scholar]
- 2.Cohnheim J. Congenitales, quergestreiftes muskelsarkon der Nireren. Virchows Arch. 1875;65:64. [Google Scholar]
- 3.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 4.Quintana E., Shackleton M., Sabel M.S., Fullen D.R., Johnson T.M., Morrison S.J. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamieson C.H., Weissman I.L., Passegué E. Chronic versus acute myelogenous leukemia: a question of self-renewal. Canc Cell. 2004;6:531–533. doi: 10.1016/j.ccr.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Dolatabadi S., Jonasson E., Lindén M., Fereydouni B., Bäcksten K., Nilsson M. JAK-STAT signalling controls cancer stem cell properties including chemotherapy resistance in myxoid liposarcoma. Int J Canc. 2019;145:435–449. doi: 10.1002/ijc.32123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inui M., Martello G., Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 8.Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 10.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 11.Ørom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 13.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 14.Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Canc Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 15.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S.Q., Jiang S., Li C., Zhang B., Li Q.J. miR-17-92 cluster targets phosphatase and tensin homology and Ikaros Family Zinc Finger 4 to promote TH17-mediated inflammation. J Biol Chem. 2014;289:12446–12456. doi: 10.1074/jbc.M114.550723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Büssing I., Slack F.J., Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Sun X., Xu C., Tang S.C., Wang J., Wang H., Wang P. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Canc Gene Ther. 2016;23:83–89. doi: 10.1038/cgt.2016.3. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z., Li Y., Kong D., Ahmad A., Banerjee S., Sarkar F.H. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Canc Lett. 2010;292:141–148. doi: 10.1016/j.canlet.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 21.Lobo N.A., Shimono Y., Qian D., Clarke M.F. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 22.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Brien C.A., Pollett A., Gallinger S., Dick J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 24.Hermann P.C., Huber S.L., Herrler T., Aicher A., Ellwart J.W., Guba M. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Takaishi S., Okumura T., Tu S., Wang S.S., Shibata W., Vigneshwaran R. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cell. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu N., Papagiannakopoulos T., Pan G., Thomson J.A., Kosik K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X., Yue Y., Wang R., Gong B., Duan Z. MicroRNA-145 inhibits tumorigenesis and invasion of cervical cancer stem cells. Int J Oncol. 2017;50:853–862. doi: 10.3892/ijo.2017.3857. [DOI] [PubMed] [Google Scholar]
- 28.Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 29.Ramachandran I., Ganapathy V., Gillies E., Fonseca I., Sureban S.M., Houchen C.W. Wnt inhibitory factor 1 suppresses cancer stemness and induces cellular senescence. Cell Death Dis. 2014;5:e1246. doi: 10.1038/cddis.2014.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chien C.S., Wang M.L., Chu P.Y., Chang Y.L., Liu W.H., Yu C.C. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Canc Res. 2015;75:2553–2565. doi: 10.1158/0008-5472.CAN-14-2215. [DOI] [PubMed] [Google Scholar]
- 31.Shell S., Park S.M., Radjabi A.R., Schickel R., Kistner E.O., Jewell D.A. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovnicki J., Gan Y., Feng T., Li Y., Xie N., Ho C.H. LIN28B promotes the development of neuroendocrine prostate cancer. J Clin Invest. 2020;130:5338–5348. doi: 10.1172/JCI135373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Canc Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Jiang P., Li J., Peng M., Zhao X., Zhang X. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37:3822–3838. doi: 10.1038/s41388-018-0237-9. [DOI] [PubMed] [Google Scholar]
- 35.Shimono Y., Zabala M., Cho R.W., Lobo N., Dalerba P., Qian D. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jian X., He H., Zhu J., Zhang Q., Zheng Z., Liang X. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Canc. 2020;19:20. doi: 10.1186/s12943-020-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song S.J., Poliseno L., Song M.S., Ala U., Webster K., Ng C. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han M., Wang S., Fritah S., Wang X., Zhou W., Yang N. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain. 2020;143:512–530. doi: 10.1093/brain/awz406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., Wu N., Liu L., Dong H., Liu X. microRNA-128-3p overexpression inhibits breast cancer stem cell characteristics through suppression of Wnt signalling pathway by down-regulating NEK2. J Cell Mol Med. 2020;24:7353–7369. doi: 10.1111/jcmm.15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Meng H., Guo K. Inhibition of MicroRNA-302c on stemness of colon cancer stem cells via the CARF/Wnt/β-Catenin Axis. Dig Dis Sci. 2020 Jul 2 doi: 10.1007/s10620-020-06435-8. [DOI] [PubMed] [Google Scholar]
- 41.El Helou R., Pinna G., Cabaud O., Wicinski J., Bhajun R., Guyon L. miR-600 acts as a bimodal switch that regulates breast cancer stem cell fate through WNT signaling. Cell Rep. 2017;18:2256–2268. doi: 10.1016/j.celrep.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 42.Hsieh I.S., Chang K.C., Tsai Y.T., Ke J.Y., Lu P.J., Lee K.H. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 43.Jiang N., Zou C., Zhu Y., Luo Y., Chen L., Lei Y. HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and Notch signaling. Theranostics. 2020;10:2553–2570. doi: 10.7150/thno.41120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin W. Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial-mesenchymal transition. Cells. 2020;9 doi: 10.3390/cells9010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su Y.J., Lai H.M., Chang Y.W., Chen G.Y., Lee J.L. Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J. 2011;30:3186–3199. doi: 10.1038/emboj.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia Q., Han T., Yang P., Wang R., Li H., Zhang J. MicroRNA-28-5p regulates liver cancer stem cell expansion via IGF-1 pathway. Stem Cell Int. 2019;2019:8734362. doi: 10.1155/2019/8734362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao C., Su L., Shan J., Zhu C., Liu L., Liu C. IGF/STAT3/NANOG/Slug signaling Axis simultaneously controls epithelial-mesenchymal transition and stemness maintenance in colorectal cancer. Stem Cell. 2016;34:820–831. doi: 10.1002/stem.2320. [DOI] [PubMed] [Google Scholar]
- 48.Liu S., Cheng K., Zhang H., Kong R., Wang S., Mao C. Methylation status of the nanog promoter determines the switch between cancer cells and cancer stem cells. Adv Sci (Weinh) 2020;7:1903035. doi: 10.1002/advs.201903035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y., Kanwar S.S., Patel B.B., Oh P.S., Nautiyal J., Sarkar F.H. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis. 2012;33:68–76. doi: 10.1093/carcin/bgr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourguignon L.Y., Earle C., Wong G., Spevak C.C., Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–160. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen M.W., Yang S.T., Chien M.H., Hua K.T., Wu C.J., Hsiao S.M. The STAT3-miRNA-92-wnt signaling pathway regulates spheroid formation and malignant progression in ovarian cancer. Canc Res. 2017;77:1955–1967. doi: 10.1158/0008-5472.CAN-16-1115. [DOI] [PubMed] [Google Scholar]
- 52.Rokavec M., Öner M.G., Li H., Jackstadt R., Jiang L., Lodygin D. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramakrishnan V., Xu B., Akers J., Nguyen T., Ma J., Dhawan S. Radiation-induced extracellular vesicle (EV) release of miR-603 promotes IGF1-mediated stem cell state in glioblastomas. EBioMed. 2020;55:102736. doi: 10.1016/j.ebiom.2020.102736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H., Cai K., Wang J., Wang X., Cheng K., Shi F. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cell. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 55.Jiang J., Li Z., Yu C., Chen M., Tian S., Sun C. MiR-1181 inhibits stem cell-like phenotypes and suppresses SOX2 and STAT3 in human pancreatic cancer. Canc Lett. 2015;356:962–970. doi: 10.1016/j.canlet.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Rinkenbaugh A.L., Baldwin A.S. The NF-κB pathway and cancer stem cells. Cells. 2016;5 doi: 10.3390/cells5020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeh D.W., Chen Y.S., Lai C.Y., Liu Y.L., Lu C.H., Lo J.F. Downregulation of COMMD1 by miR-205 promotes a positive feedback loop for amplifying inflammatory- and stemness-associated properties of cancer cells. Cell Death Differ. 2016;23:841–852. doi: 10.1038/cdd.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li M., Pan M., You C., Zhao F., Wu D., Guo M. MiR-7 reduces the BCSC subset by inhibiting XIST to modulate the miR-92b/Slug/ESA axis and inhibit tumor growth. Breast Cancer Res. 2020;22:26. doi: 10.1186/s13058-020-01264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chikuda J., Otsuka K., Shimomura I., Ito K., Miyazaki H., Takahashi R.U. CD44s induces miR-629-3p expression in association with cisplatin resistance in head and neck cancer cells. Cancers (Basel) 2020;12 doi: 10.3390/cancers12040856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bai R., Dou K., Wu Y., Ma Y., Sun J. The NF-κB modulated miR-194-5p/IGF1R/PPFIBP axis is crucial for the tumorigenesis of ovarian cancer. J Canc. 2020;11:3433–3445. doi: 10.7150/jca.40604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Canc. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou J., Wang H., Che J., Xu L., Yang W., Li Y. Silencing of microRNA-135b inhibits invasion, migration, and stemness of CD24(+)CD44(+) pancreatic cancer stem cells through JADE-1-dependent AKT/mTOR pathway. Canc Cell Int. 2020;20:134. doi: 10.1186/s12935-020-01210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bahena-Ocampo I., Espinosa M., Ceballos-Cancino G., Lizarraga F., Campos-Arroyo D., Schwarz A. miR-10b expression in breast cancer stem cells supports self-renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep. 2016;17:648–658. doi: 10.15252/embr.201540678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng L., Zhang Y., Liu Y., Zhou M., Lu Y., Yuan L. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med. 2015;13:252. doi: 10.1186/s12967-015-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez-Bertoni H., Kotchetkov I.S., Mihelson N., Lal B., Rui Y., Ames H. A sox2:miR-486-5p Axis regulates survival of GBM cells by inhibiting tumor suppressor networks. Canc Res. 2020;80:1644–1655. doi: 10.1158/0008-5472.CAN-19-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao L., Guo Q., Li X., Yang X., Ni H., Wang T. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMed. 2019;41:395–407. doi: 10.1016/j.ebiom.2019.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J., Li M., Han X., Wang H., Wang X., Ma G. MiR-1976 knockdown promotes epithelial-mesenchymal transition and cancer stem cell properties inducing triple-negative breast cancer metastasis. Cell Death Dis. 2020;11:500. doi: 10.1038/s41419-020-2711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meurette O., Mehlen P. Notch signaling in the tumor microenvironment. Canc Cell. 2018;34:536–548. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 69.Nwaeburu C.C., Abukiwan A., Zhao Z., Herr I. Quercetin-induced miR-200b-3p regulates the mode of self-renewing divisions in pancreatic cancer. Mol Canc. 2017;16:23. doi: 10.1186/s12943-017-0589-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Huang G., Wang M., Li X., Wu J., Chen S., Du N. TUSC7 suppression of Notch activation through sponging MiR-146 recapitulated the asymmetric cell division in lung adenocarcinoma stem cells. Life Sci. 2019;232:116630. doi: 10.1016/j.lfs.2019.116630. [DOI] [PubMed] [Google Scholar]
- 71.Kwon H., Song K., Han C., Zhang J., Lu L., Chen W. Epigenetic silencing of miRNA-34a in human cholangiocarcinoma via EZH2 and DNA methylation: impact on regulation of notch pathway. Am J Pathol. 2017;187:2288–2299. doi: 10.1016/j.ajpath.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kang L., Mao J., Tao Y., Song B., Ma W., Lu Y. MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway. Canc Sci. 2015;106:700–708. doi: 10.1111/cas.12656. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Wang S., Cai L., Zhang F., Shang X., Xiao R., Zhou H. Inhibition of EZH2 attenuates sorafenib resistance by targeting NOTCH1 activation-dependent liver cancer stem cells via NOTCH1-related MicroRNAs in hepatocellular carcinoma. Transl Oncol. 2020;13:100741. doi: 10.1016/j.tranon.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xin L., Liu L., Liu C., Zhou L.Q., Zhou Q., Yuan Y.W. DNA-methylation-mediated silencing of miR-7-5p promotes gastric cancer stem cell invasion via increasing Smo and Hes1. J Cell Physiol. 2020;235:2643–2654. doi: 10.1002/jcp.29168. [DOI] [PubMed] [Google Scholar]
- 75.Jin Y., Wang M., Hu H., Huang Q., Chen Y., Wang G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int J Biol Macromol. 2018;117:445–453. doi: 10.1016/j.ijbiomac.2018.05.151. [DOI] [PubMed] [Google Scholar]
- 76.Jeong J.Y., Kang H., Kim T.H., Kim G., Heo J.H., Kwon A.Y. MicroRNA-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Canc Lett. 2017;386:168–178. doi: 10.1016/j.canlet.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 77.Wang X., Meng Q., Qiao W., Ma R., Ju W., Hu J. miR-181b/Notch2 overcome chemoresistance by regulating cancer stem cell-like properties in NSCLC. Stem Cell Res Ther. 2018;9:327. doi: 10.1186/s13287-018-1072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brabletz S., Bajdak K., Meidhof S., Burk U., Niedermann G., Firat E. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H., Li F. Exosomes from BM-MSCs increase the population of CSCs via transfer of miR-142-3p. Br J Canc. 2018;119:744–755. doi: 10.1038/s41416-018-0254-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y.A., Lu C.Y., Cheng T.Y., Pan S.H., Chen H.F., Chang N.S. WW domain-containing proteins YAP and TAZ in the Hippo pathway as key regulators in stemness maintenance, tissue homeostasis, and tumorigenesis. Front Oncol. 2019;9:60. doi: 10.3389/fonc.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu T., Yang Y., Li Z., Lu S. MicroRNA-214-3p inhibits the stem-like properties of lung squamous cell cancer by targeting YAP1. Canc Cell Int. 2020;20:413. doi: 10.1186/s12935-020-01506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muñoz-Galván S., Felipe-Abrio B., Verdugo-Sivianes E.M., Perez M., Jiménez-García M.P., Suarez-Martinez E. Downregulation of MYPT1 increases tumor resistance in ovarian cancer by targeting the Hippo pathway and increasing the stemness. Mol Canc. 2020;19:7. doi: 10.1186/s12943-020-1130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mori M., Triboulet R., Mohseni M., Schlegelmilch K., Shrestha K., Camargo F.D. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takahashi R.U., Miyazaki H., Ochiya T. The role of microRNAs in the regulation of cancer stem cells. Front Genet. 2014;4:295. doi: 10.3389/fgene.2013.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 86.Takahashi R.U., Prieto-Vila M., Kohama I., Ochiya T. Development of miRNA-based therapeutic approaches for cancer patients. Canc Sci. 2019;110:1140–1147. doi: 10.1111/cas.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yáñez-Mó M., Siljander P.R., Andreu Z., Zavec A.B., Borràs F.E., Buzas E.I. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Usman W.M., Pham T.C., Kwok Y.Y., Vu L.T., Ma V., Peng B. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9:2359. doi: 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]