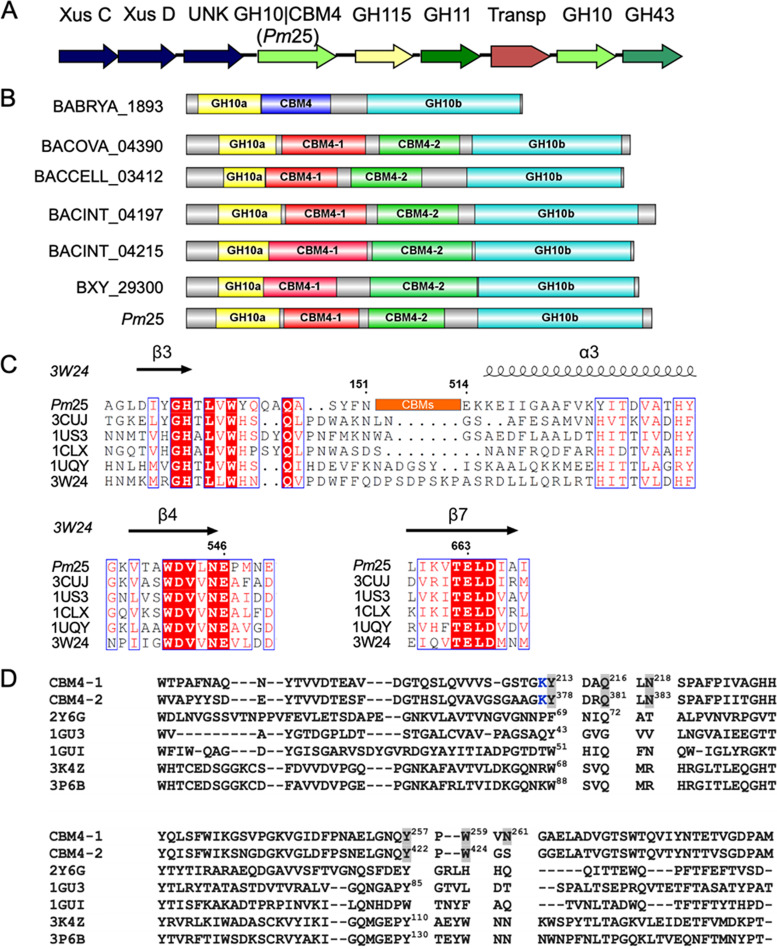
FIG 1.
Modular architecture of Xyn10C-like protein. (A) Schematic representation of the putative xylan-active gene cluster. (B) Insertional architecture of Xyn10C-like proteins; the length of the amino acid is proportional to the length of the colored box. Shown are BABRYA_1893 (29), BACOVA_04390 (71), BACCELL_03412 (72), BACINT_04197 and BACINT_04215 (33), BXY_29300 (30), and Pm25 (17). (C) Amino acid alignment of Pm25 with other native GH10 xylanases, designated by their PDB codes: 3CUJ, Cellulomonas fimi xylanase/cellulase Cex (CfXyn10A); 1US3, Cellvibrio japonicus xylanase 10C; 1CLX, Pseudomonas fluorescens xylanase A; 1UQY, Cellvibrio mixtus xylanase B; 3W24, Thermoanaerobacterium saccharolyticum xylanase A. The β strands and α helices are shown above the aligned sequences. Numbering corresponds to the sequences of Pm25. Residues in red background are conserved, and those within blue frames are conservatively substituted. (D) Amino acid alignment of CBM4s in Pm25 with well-characterized CBM4s in the literature. CBM4-1 is the first CBM4 in Pm25; CBM4-2 is the second CBM4 in Pm25. PDB 2Y6G, CBM4 from Rhodothermus marinus xylanase; 1GU3, CBM4 from Cellulomonas fimi endoglucanase C; 1GUI, CBM4 from Thermotoga maritima laminarinase 16A; 3K4Z, CBM4 from Clostridium thermocellum cellulase CbhA; 3P6B, CBM4 from Clostridium thermocellum cellulase CelK. Residues highlighted in gray were used for mutagenesis. Numbered residues in PDB sequences indicate that they are responsible for ligand binding according to the literature. The K in blue is potentially labeled with N-hydroxysuccinimide dye, which was described in Materials and Methods for the MST experiment.