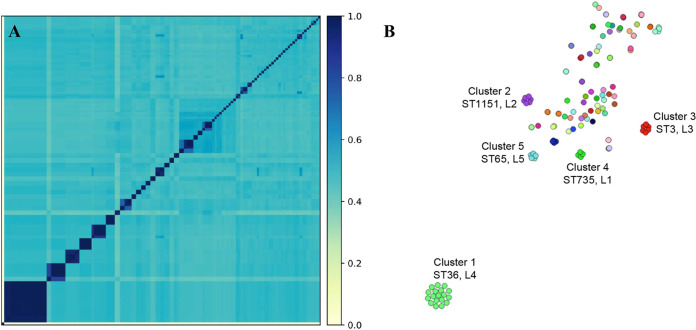
FIG 4.
Vibrio parahaemolyticus genome relatedness using whole-genome distance comparisons. (A) Genome distance using k-mers with an all-against-all comparison. (B) Genomically related isolate network using PopPUNK to demonstrate specific genomic clusters that are epidemiologically related within a cluster but genetically distinct between the groups. Colors represent the same genomic group. The distance between groups indicates relatedness as determined using whole-genome distance. Clusters are labeled with the unique cluster identification from this analysis (clusters 1 to 5), the ST that comprises the cluster, and the likelihood tree partition to which the isolates belong (“LX,” where “X” is the partition number in Fig. 3).