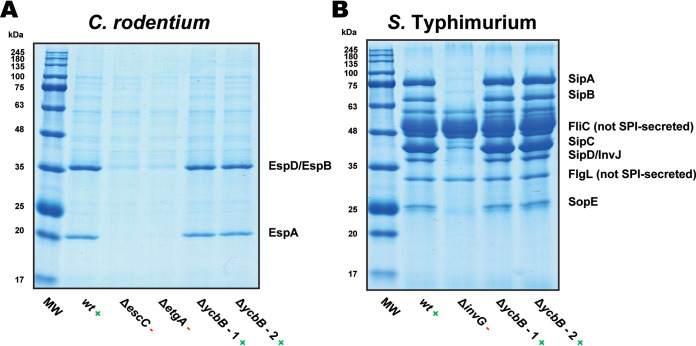
FIG 4.
Secreted-protein profiles from ΔycbB mutants of C. rodentium and S. Typhimurium. (A) Secreted-protein profiles from C. rodentium strain DBS100, which utilizes a LEE (locus of enterocyte effacement)-encoded T3SS. The wild-type (wt) strain was used as a positive control, and ΔescC and ΔetgA mutant strains were used as negative controls. The ΔycbB strain is seen to retain LEE T3SS activity. (B) Secreted-protein profiles from S. Typhimurium strain SL1344, which utilizes an SPI-1 (Salmonella pathogenicity island 1)-encoded T3SS. The wild-type (wt) strain was used as a positive control, and the ΔinvG mutant strain was used as a negative control. The ΔycbB strain is seen to retain SPI-1 T3SS activity. The two ΔycbB lanes in each of panels A and B represent two independent isolates, which should be genetically identical. Lanes MW are molecular weight markers.