Figure EV2. Gene silencing of Lemd2, Vps4a and Vps4b in primary neuronal cultures.
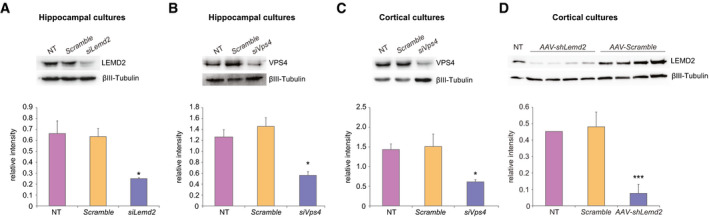
- Gene silencing of Lemd2 in hippocampal cultures by siRNA. Upper panel: Representative Western blot for LEMD2 protein in primary hippocampal neurons, NT (non‐transfected), Scramble (transfected with control siRNA), siLemd2 (transfected with siRNA against Lemd2), Lower panel: Western blot quantification of LEMD2 in hippocampal cultures, n = 4 independent primary cultures. One‐way Welch’s ANOVA followed by Tukey post hoc test, F 2,9 = 16.783, P = 0.011, NT vs siLemd2 P = 0.013, Scramble vs siLemd2 P = 0.019. Data are presented as mean ± SEM, *P < 0.05 compared to NT.
- Gene silencing of Vps4 in hippocampal cultures by siRNA. Upper panel: Representative Western blot for VPS4 protein level in primary hippocampal neurons. NT (non‐transfected), Scramble (transfected with control siRNA), siVps4 (transfected with siRNAs against Vps4a and Vps4b). Lower panel: Western blot quantification of VPS4 protein level in primary hippocampal cultures, n = 3 independent experiments, one‐way Welch’s ANOVA followed by Tukey post hoc test, F 2,6 = 14.226, P = 0.027, NT vs siVps4 P = 0.063, Scramble vs siLemd2 P = 0.045. Data are presented as mean ± SEM, *P < 0.05 compared to scramble.
- Gene silencing of Vps4 in cortical cultures by siRNA. Upper panel: Representative Western blot for VPS4 protein level in primary cortical neurons. NT (non‐transfected), scramble (transfected with control siRNA), Vps4 (transfected with siRNA against Vps4a and Vps4b), Lower panel: Western blot quantification of VPS4 protein level in primary cortical cultures, n = 3 independent experiments, one‐way ANOVA followed by Tukey post hoc test, F 2,8 = 9.423, P = 0.014, NT vs siVps4 P = 0.041, scramble vs siVps4 P = 0.015. Data are presented as mean ± SEM, *P < 0.05 compared to NT.
- Gene silencing of Lemd2 in cortical cultures by shRNA. Upper panel: Western blot for LEMD2 in primary cortical neurons, NT (non‐transfected), Scramble (transfected with control shRNA), AAV‐shLemd2 (transfected with shRNA against Lemd2). Lower panel: Western blot quantification of LEMD2 protein level, n = 4 independent experiments, unpaired t‐test, t(6) = 5.98, P = 0.000979. Data are presented as mean ± SEM, ***P < 0.001 compared to Scramble.
Source data are available online for this figure.
