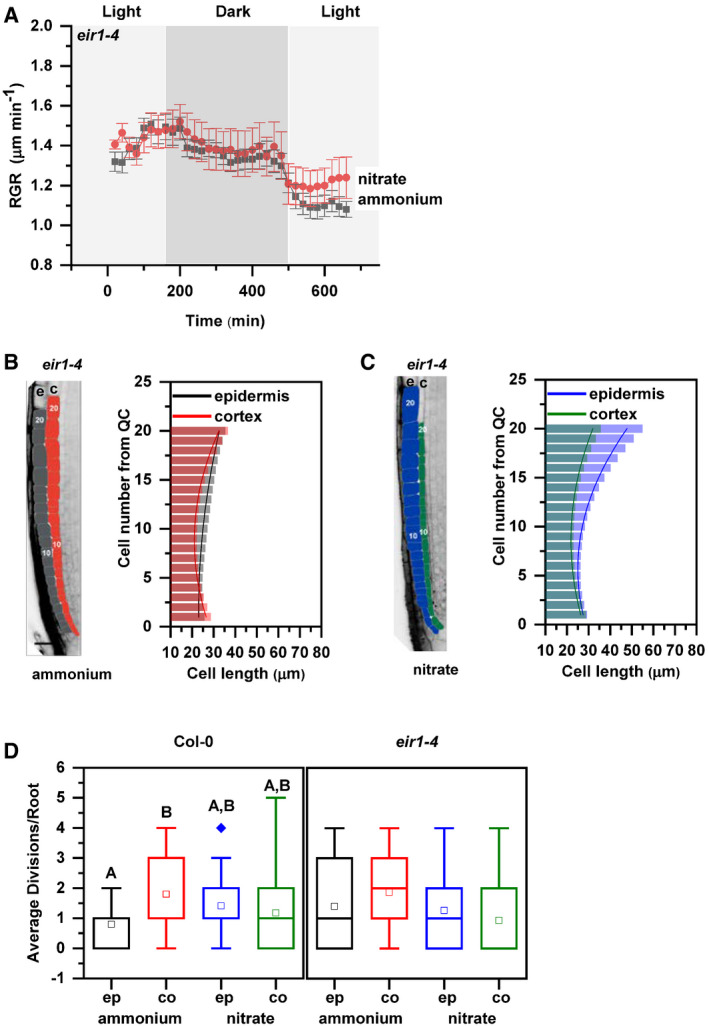
Figure 3. Primary root growth kinetics of eir1‐4 roots transferred to ammonium or nitrate amended medium.
-
ARoot growth rate (RGR in µm/min) of eir1‐4 roots transferred 5 DAG to ammonium (gray) or nitrate (red) containing medium over a period of 680 min. Data represent the geometric mean (± standard error, SE) of 3 biological replicates (n = 5 roots/condition). Light and dark periods are highlighted in light or dark gray.
-
B, CRepresentation and quantification of cell length in epidermal (e) and cortical (c) cell files. Optical, longitudinal sections of 5 DAG eir1‐4 roots 12 HAT to ammonium (B) or nitrate (C) supplemented media. The first 20‐20 epidermal and cortex cells (from quiescent center (QC)) are highlighted in gray and in red on ammonium (B) and in blue and green on nitrate (C), respectively. Scale bar = 30 µm. Column bars denote the geometric mean of cell length at the respective positions. Lines represent a polynomial regression fit, with calculated slopes between cells 10 and 20 of 0.75884 ± 0.02624 (ammonium, epidermis), 1.13088 ± 0.08446 (ammonium, cortex) and 2.06912 ± 0.10341 (nitrate, epidermis), 0.99878 ± 0.07278 (nitrate, cortex). Data are derived from 3 biological replicates, n = 9 (ammonium) and 8 (nitrate) roots.
-
DAverage number of cell divisions along the epidermis (ep) and cortex (co) in 5 DAG old Col‐0 and eir1‐4 root tips 12 HAT to ammonium or nitrate supplemented media. Data are derived from n = 15 and n = 17 roots of Col and n = 10 and n = 9 roots of eir1‐4 on ammonium and nitrate, respectively. Statistical significance was evaluated with ANOVA at P < 0.05. The box chart components are defined as; box (25–75%), central band (median line), central box (mean), and the range is within 1.5IQR.
Source data are available online for this figure.
