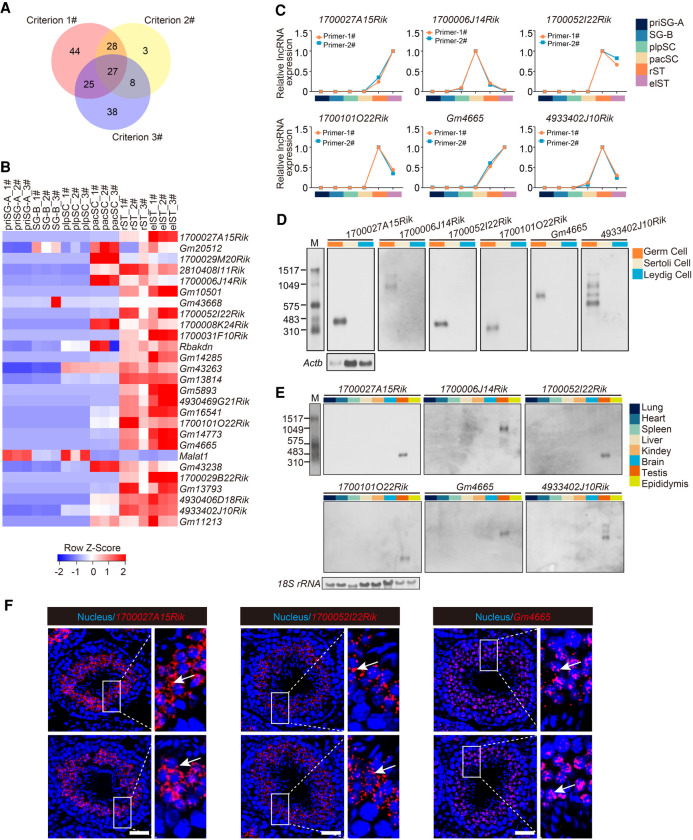
Figure 2.
Characterization of potential functional lncRNAs in mouse spermatogenesis. (A) Venn diagram showing the number of lncRNAs meeting the three screening criteria to determine potential functional lncRNAs. (B) Heat map showing relative expression level of the 27 candidate lncRNAs in each germ cell type. 1#, 2#, and 3# refer to three biological replicates per germ cell type. (C) Expression validation of six candidate lncRNAs in each germ cell type by qRT-PCR. Two independent pairs of primers were used for each candidate lncRNA (primer-1# and primer-2#); also see Supplemental Figure S3A. (D) Northern blotting analysis of six candidate lncRNAs in germ cells, Sertoli cells, and Leydig cells. Actb mRNA served as a loading control. (M) RNA molecular weight marker. (E) Northern blotting analysis of six candidate lncRNAs in multiple tissues. 18S rRNA served as a loading control. (M) RNA molecular weight marker. (F) Subcellular localization of candidate lncRNAs (red) from testis sections via RNAscope analysis. Nuclei were stained with DAPI (blue). Representative images with an enlargement of the white framed region are shown on the right. Scale bar, 50 μm.