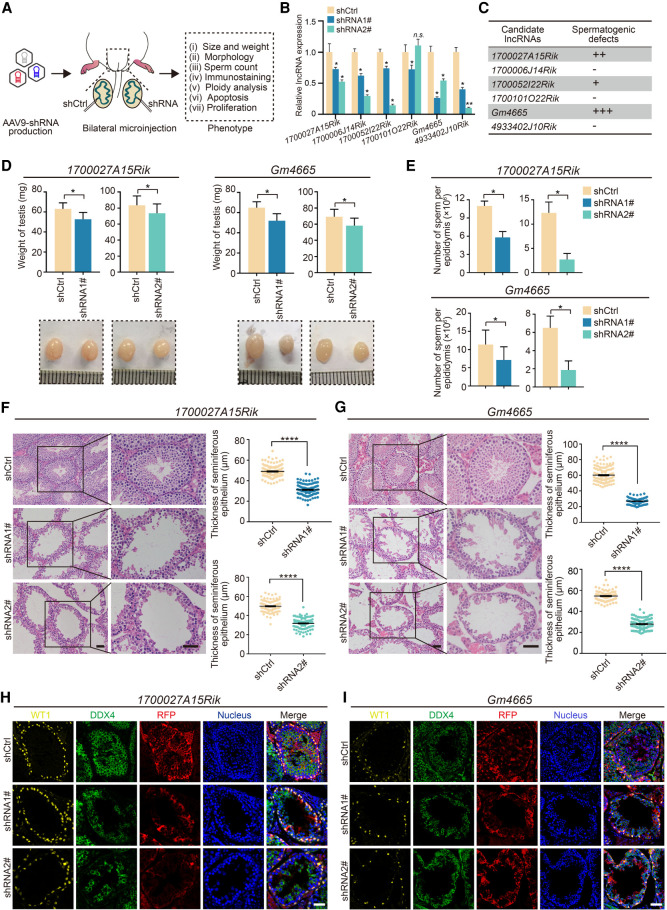
Figure 3.
Functional screening of candidate lncRNAs in mouse spermatogenesis in vivo. (A) Schematic overview of in vivo functional screening strategy. (B) qRT-PCR analysis of the efficacy of lncRNAs knockdown in vivo by AAV9-shRNA-RFP. Data represent the mean ± SEM for three biological replicates. (*) P < 0.05, (n.s.) P > 0.05 compared with shCtrl, Student's t-test. (C) Spermatogenesis phenotype profiles following lncRNAs knockdown. All results in this figure were collected 4 wk after microinjection with AAV9. (D) Testis morphology and weight from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice. (Top) The average weight of testes; (bottom) representative images of testes. Data represent mean ± SEM (n = 5). (*) P < 0.05 compared with shCtrl, Student's t-test. (E) Sperm counts in the cauda epididymis from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice. Data represent mean ± SEM (n = 5). (*) P < 0.05 compared with shCtrl, Student's t-test. (F,G) H&E staining of testis sections from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice. (Left) Representative staining images; (right) thickness of seminiferous epithelium. Data represent mean ± SEM of at least 100 seminiferous tubules from three mice. (****) P < 0.0001 compared with shCtrl, Student's t-test. Scale bar, 50 μm. (H,I) Immunostaining of testis sections from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice for WT1 (yellow), DDX4 (green), and RFP (red). Nuclei were stained with DAPI (blue). Scale bar, 50 μm.