Abstract
As the most complex organ of the human body, the brain is composed of diverse regions, each consisting of distinct cell types and their respective cellular interactions. Human brain development involves a finely tuned cascade of interactive events. These include spatiotemporal gene expression changes and dynamic alterations in cell-type composition. However, our understanding of this process is still largely incomplete owing to the difficulty of brain spatiotemporal transcriptome collection. In this study, we developed a tensor-based approach to impute gene expression on a transcriptome-wide level. After rigorous computational benchmarking, we applied our approach to infer missing data points in the widely used BrainSpan resource and completed the entire grid of spatiotemporal transcriptomics. Next, we conducted deconvolutional analyses to comprehensively characterize major cell-type dynamics across the entire BrainSpan resource to estimate the cellular temporal changes and distinct neocortical areas across development. Moreover, integration of these results with GWAS summary statistics for 13 brain-associated traits revealed multiple novel trait–cell-type associations and trait-spatiotemporal relationships. In summary, our imputed BrainSpan transcriptomic data provide a valuable resource for the research community and our findings help further studies of the transcriptional and cellular dynamics of the human brain and related diseases.
The brain is the most complex organ in the human body (Bassett and Gazzaniga 2011). The past decade has witnessed a tremendous progress toward a deep understanding of transcriptional regulation of the developing human brain (Darmanis et al. 2015). Brain development involves precisely tuned gene expression dynamics with strong spatiotemporal specificity and alterations of many cell types. For example, neuronal development is generally summarized as neurogenesis, neuronal migration, and axon guidance. During this process, neurons undergo extreme morphological and functional changes, combined with the formation of synapses and intricate neural circuits (Miller et al. 2014; Weyn-Vanhentenryck et al. 2018), to build connections between neurons. Abnormalities during brain development have been related to various neurodevelopmental diseases, including psychiatric disorders (Peralta and Cuesta 2017). Specific brain regions or developmental stages were found to be involved in different diseases. For a few examples, autism spectrum disorder (ASD)–related and intellectual disability (ID)–related genes tended to be highly expressed during pre- and perinatal stages (Courchesne et al. 2007; D'Haene et al. 2016); distinguished patterns of gene expression between frontal and temporal cortices were reported to be significantly attenuated in brains of autism patients (Voineagu et al. 2011); schizophrenia (SCZ)-related genes were found highly expressed during prenatal development (Gilman et al. 2012); and several regions, including the amygdala, hippocampus, and prefrontal cortex, were found to be related to bipolar disorder (Ellison-Wright and Bullmore 2010; Chai et al. 2011). Although the evidence is accumulating, such studies are still very limited on the highly qualified brain gene expression data linking to the neurodevelopmental disease. To better understand the role of disease genes during development, there is a pressing need for a comprehensive and a complete annotation of the developing brain.
So far, two large resources, the BrainSpan Atlas of the Developing Human Brain (Kang et al. 2011) and BrainCloud (Colantuoni et al. 2011), have provided comprehensive transcriptomic data for human brain development. Many studies, including two of ours (Jia et al. 2017, 2018), used the BrainSpan data to study disease-associated genes. BrainSpan contains expression data across 26 brain regions ranging in age from 8 postconceptual weeks (pcw) to 40 yr, covering the entire developmental process. Critically, because human brain samples are extremely hard to collect (Farahany et al. 2018), not all regions have been profiled across all time points, and there is a high level of incompleteness. Nearly ∼80% of the individuals profiled in BrainSpan had at least one missing transcriptome. Such missing data problems are prevalent in biological studies and are even worse in human brain research, where it is practically impossible to solve. For example, the Genotype-Tissue Expression (GTEx) Consortium (The GTEx Consortium. 2015) collected 14,787 transcriptomes from 948 patients spanning 53 tissues, including 13 brain regions (version 7). However, all these samples were from adult brains (age ≥ 20), and the sample size for the brain regions was among the smallest compared with the other tissues profiled in GTEx. Therefore, computational approaches to impute transcriptome-wide gene expression data leading to a complete and reference-driven spatiotemporal data grid have become necessary. So far, several transcriptomic imputation approaches by integrating eQTL and genome-wide association studies (GWAS) summary data have been applied to transcriptomic imputation (Huckins et al. 2019; Zhang et al. 2019a). However, these imputation methods rely on tissue-specific (or brain region–specific) eQTL information (Wang et al. 2016; Xu et al. 2020). Although the BrainSpan Atlas is the most comprehensive transcriptomic data collection for human brain development, we did not find any eQTL data in the BrainSpan consortium with spatiotemporal specificity. On the other hand, a few tensor-based works have been shown to successfully impute epigenomic data at the genome-wide level (Durham et al. 2018); however, these approaches have not been applied to transcriptomics data yet. In this study, by considering the special temporal and spatial design of BrainSpan, we developed a computational approach to leverage the natural tensor structure of the data to perform transcriptome-wide imputation. We transformed all transcriptome data into a three-dimensional (3D) tensor, consisting of spatial, temporal, and gene expression components. This tensor is first compressed into a low-rank representation of the data and subsequently transformed to impute entire transcriptome-wide expression profiles.
To understand the neurodevelopmental process, it is crucial to characterize how cell-type composition (CTC) changes across different regions and developmental stages. For >20 yr, researchers have classified neurons that populate the neocortex into two major classes: projection neurons (excitatory) and GABAergic interneurons (inhibitory) (Parnavelas 2000). Transcriptional profiling of these neurons, however, has not been made possible until the recent advances in single-cell RNA sequencing (scRNA-seq) technologies (Darmanis et al. 2015; Lake et al. 2016). scRNA-seq of brain samples has characterized a number of cell types, including their respective marker genes (Lake et al. 2016). Further augmented by the recent advances in machine learning, several deconvolution methods have been developed to reliably infer CTC from bulk gene expression data (Newman et al. 2015; Aran et al. 2017; Glastonbury et al. 2019).
In this work, we comprehensively characterized CTCs across the entire spatiotemporal grid of the BrainSpan resource. We subsequently integrated these imputed results with GWAS summary statistics covering 13 brain-associated traits, aiming to uncover novel links between cell types, brain regions, developmental stages, and brain-related phenotypes, including major psychiatric disorders.
Results
Overview of workflow
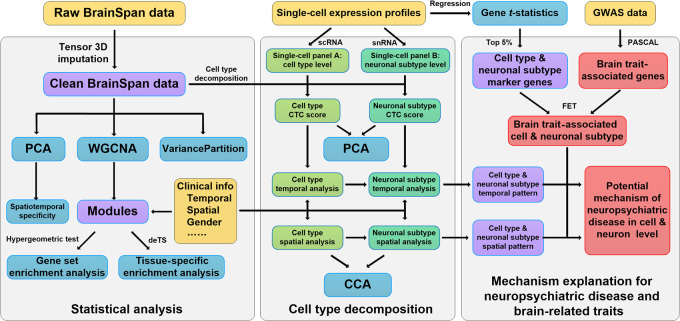
Our goal is to decode CTCs in spatiotemporal specificity throughout human brain development and then uncover disease-relevant cell types for major brain disorders. To this end, we collected various public data sources and conducted a range of bioinformatics and functional genomic analyses (Fig. 1). We constructed comprehensive resources for brain, including both bulk (Miller et al. 2014) and single-cell (Newman et al. 2015; Aran et al. 2017) expression profiles (Fig. 1). Different analytical strategies were applied for each data set. For gene expression data from BrainSpan, we completed the spatiotemporal data grid using a tensor-based imputation strategy, generating a valuable resource for the research community of brain-related studies. We applied weighted gene coexpression network analysis (WGCNA) (Langfelder and Horvath 2008) to study the temporal and spatial specificity of human brain expression profiles. We collected two single-cell expression studies, each with particular focus on brain cell types and neurons during development and subsequently used these data as reference panels for cell-type deconvolution (Newman et al. 2015) of the bulk expression data from BrainSpan, for example, characterizing the dynamic changes of brain CTC during development, and also investigated cell types in which the disease-associated genes were enriched.
Figure 1.
Analysis workflow. Details are provided in the Methods section. Color of the boxes is as follows: yellow, external input data; blue, statistical analysis methods; green, cell-type composition (CTC) result; purple, intermediate analysis result; and red, GWAS-related analysis.
Robust imputation completes the BrainSpan transcriptome data
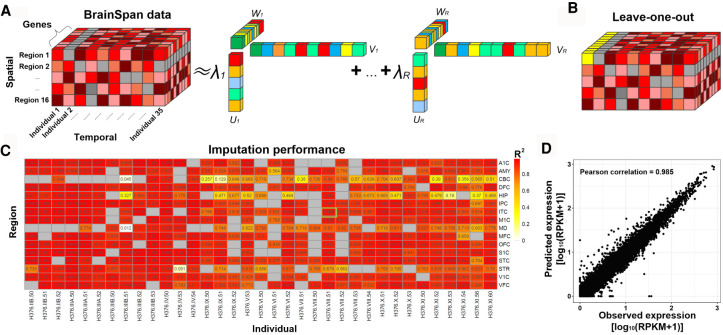
To infer missing data points in the spatiotemporal data grid, the original BrainSpan data was transformed into a 3D tensor (18,911 × 42 × 26) (Fig. 2A). In this original tensor, some individuals and brain regions had high missing data rates. For example, the individual H376.V.50-52 had only three data points and a missing data rate of 23/26; the region temporal neocortex (TCx) had only one data point, with a missing rate of 41/42. Our initial application of the CANDECOMP/PARAFAC (CP) algorithm showed that these individuals or regions tended to have a low R2-values (<0.6 for both cases mentioned above), indicating a low imputation performance. Based on this initial evaluation, we excluded individuals or regions with a missing data rate >50%, resulting in a working tensor (18,911 × 35 × 16) with 478 measured and 82 missing transcriptomes to be imputed. By applying the CP method to this working tensor, we evaluated the performance of our imputation strategy using leave-one-out (LOO) cross-validation. As shown in Figure 2, B through D, and Supplemental Figure S1, the Pearson correlation coefficient (PCC) values from LOO ranged from 0.11–0.99 (median value: 0.96) and R2 ranged from 0.012–0.98 (median: 0.93). To avoid potential high correlation confounded by the mean gene expression level, we also evaluated the performance between the imputed results and the observed expression for each sample itself (i.e., the matched pair), as well as the performance between the imputed results with all the other samples (i.e., the unmatched pairs). As shown in Supplemental Figure S2, A through C, the PCC and R2 between imputed results with the original data (the matched pair) are significantly higher than the unmatched pairs’ gene expression profiles and incorrect pairs. Additionally, we assessed the correlation across samples for each gene. Based on the gene abundance level, we divided all genes into three groups, including low-, moderate-, and high-abundance groups. As shown in Supplemental Figure S2D, the median PCC for low-, moderate-, and high-abundance gene groups is 0.07, 0.62, and 0.71, respectively, indicating that the performance is relatively better for the moderate and high group than the low-abundance group. Overall, this performance showing that our approach robustly imputed the entire grid of spatiotemporal transcriptomes in BrainSpan.
Figure 2.
BrainSpan data completion and imputation evaluation. (A) CANDECOMP/PARAFAC (CP) tensor decomposition framework. (B) The schematic diagram of leave-one-out (LOO) imputation evaluation strategy. Gray boxes represent missing values. Yellow boxes represent one leave-out sample, assuming all gene expression values in that column are not available. X-axis, y-axis, and z-axis correspond to individuals, brain regions, and genes, respectively. (C) R2 of imputation performance of BrainSpan data sets by LOO strategy. (D) One example showing the correlation between observed and predicted gene expression.
Differences between individuals and temporal stages drive specific gene expression
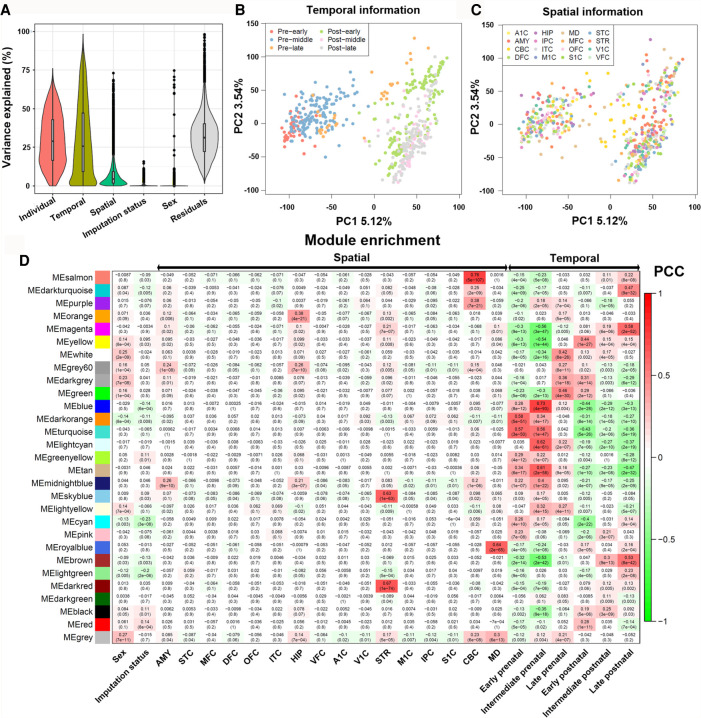
To identify biological or technical factors that contribute to the variance in gene expression, we applied variancePartition (Hoffman and Schadt 2016) to the spatiotemporal grid of gene expression profiles and estimated how much of the variance could be explained by each factor (Fig. 3A). For each gene, we calculated the percentage of its variation attributable to individuals (35 patients), developmental stages (six stages), brain regions (16 regions), imputation status (imputed or measured), and sex (male/female). The remaining, unexplained variance in expression was termed residual variation. As shown in Fig. 3A, the source of individuals explained the largest component (median value: 30%). Developmental stages and brain regions were the second and third most contributive factors, explaining a median of 18.3% and 4.3% of the expression variance, respectively. Because the factors individual and stage were correlated, we further conducted an independent analysis (Supplemental Fig. S3). The result showed that the individual could explain a median of 61.0%, whereas the stage could explain 35.1% variance. In contrast, the factors sex and imputation status could only explain a very small proportion of the variance, with their median values being <0.1% (Fig. 3A). This result further confirmed that our imputation results were robust with respect to the original expression distribution and did not create much batch effect between the measured and imputed transcriptomes. In addition, we conducted a principal component analysis (PCA) and compared each of these factors with the first two principal components (PCs). As shown in Figure 3B, early prenatal samples and late postnatal samples were clearly separable (largely by PC1), whereas other factors such as temporal stage (Fig. 3C), sex, and imputation status could not effectively separate the samples. Finally, the high percentage of residual variation unexplained by the factors we considered here suggested that there were other uncharacterized sources of expression variation.
Figure 3.
Gene expression in BrainSpan is influenced by spatiotemporal factors. (A) Violin plots of the percentage of variance explained by each variable over all the genes. Principal component analysis (PCA) of gene expression data from BrainSpan, where samples are colored according to their temporal (B) and spatial (C) attributes. (D) Correlation between identified modules and spatiotemporal factor indicated by WGCNA.
Coexpression analysis links gene modules to specific tissues in GTEx
We further explored gene function by performing WGCNA. We identified a total of 29 modules, each labeled with a color by following the WGCNA naming system. For each module, we next conducted association tests with the biological or technical factors mentioned above. As shown in Figure 3D, using the thresholds PCC > 0.5 and P < 1 × 10−10, eight modules were significantly associated with at least one developmental stage and four modules with brain region, respectively. No module was significantly associated with sex (Fig. 3D). Only one module was marginally associated with the imputation status (P = 8 × 10−4), further confirming that the imputation did not introduce systematic biases.
We also conducted a tissue-specific enrichment analysis (TSEA) using GTEx data. We applied our recently developed tool, deTS, and examined if any module was enriched for tissue-specific genes. GTEx has 13 brain regions, all of which were from adults (age ≥ 20). Because these modules were obtained using transcriptome data of brain tissues, the TSEA results served as an independent validation for our imputation. As expected, brain tissues were the most strongly enriched tissues for these modules (Supplemental Fig. S4). In addition, modules that were associated with certain brain regions as defined by the original BrainSpan annotation were found to be associated with similar brain regions in GTEx. For example, the salmon module was enriched in the cerebellar cortex (CBC; P = 5 × 10−107) as defined by BrainSpan annotations. Correspondingly, the salmon module was associated with GTEx cerebellum (P = 5 × 10−74) and cerebellar hemisphere (P = 2.0 × 10−70). The dark red module was enriched in the striatum (STR) (P = 1 × 10−74) in BrainSpan and in the basal ganglia (putamen [P = 3 × 10−36], caudate [P = 2 × 10−33], and nucleus accumbens [P = 4 × 10−32]) in GTEx. The full names of each brain region were summarized in Supplemental Table S1. Only modules that were associated with late developmental stages from BrainSpan (brown and magenta) were found associated with GTEx brain tissues. This was in line with the fact that GTEx brain tissues were all >20 yr old. Modules associated with prenatal stages (tan, blue, and turquoise) were enriched for the ovary and uterus. This was likely because of the interaction of the microenvironment of the female reproductive system with fetal brain development. Collectively, TSEA and the GTEx data validated the completed BrainSpan developmental transcriptomes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched analysis of each module can be found in Supplemental Table S2.
Deconvolution analysis reveals CTCs
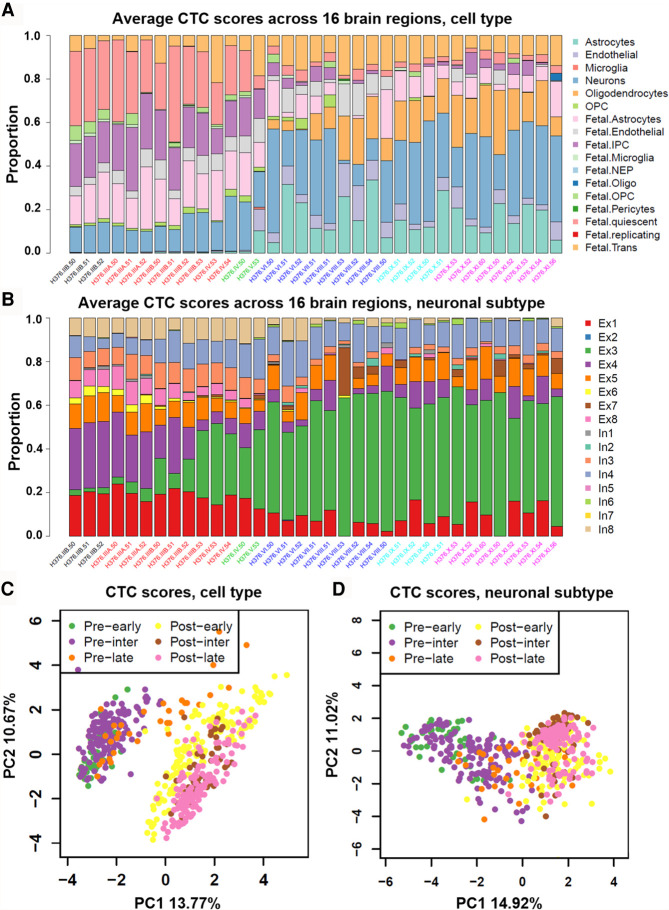
Given that we did not observe any significant associations between coexpression modules and specific cerebral areas and given that the bulk RNA-seq data represented a mixture of numerous cell types, we performed cell-type deconvolution analysis. More precisely, we applied CIBERSORT (Newman et al. 2015) in the relative mode to estimate CTC scores for each BrainSpan sample (Supplemental Table S3). To provide unique yet complementary insights, we used two reference panels for the deconvolution analysis. Panel A contained cell types from both fetal and adult brains, whereas panel B contained different neurons from an adult brain. For improved presentation, we took the average CTC scores across 16 regions for each individual to visualize the composition of cell types (panel A) (Fig. 4A) and neuronal subtypes (panel B) (Fig. 4B), respectively. PCA revealed that the largest contribution to the variance in the composition of cell types or neuronal subtypes was explained by differences between prenatal and postnatal samples (Fig. 4C,D). Other factors, such as sex and imputation status, did not show significant differences in CTC scores. However, CIBERSORT estimates the relative composition for each cell type, and thus, the CIBERSORT results need to be interpreted carefully. For example, although adult cell types are not expected in fetal samples, CIBERSORT still reported a small fraction of adult cell types because these cells are included in the reference panel. We applied CIBORSORT twice using all cell types of panel A and using only the adult cell types of panel A, respectively. As shown in Supplemental Figure S5, our results revealed that the CTC scores of most cell types had strong correlation (PCC > 0.9). These results showed the robustness of the “relative score.”
Figure 4.
Deconvolution analysis of BrainSpan bulk-cell RNA sequencing data. Schematic diagram of relative percent (16 regions averaged) of cell composition by two single-cell panels: (A) panel A, six adult + 11 fetal brain-related cell types, and (B) panel B, eight excitatory and eight inhibitory neuronal subtypes. Samples are colored according to their temporal attributes. PCA plot analysis according to CTC scores in panel A (C) and panel B (D).
CTC changes across temporal stage and region
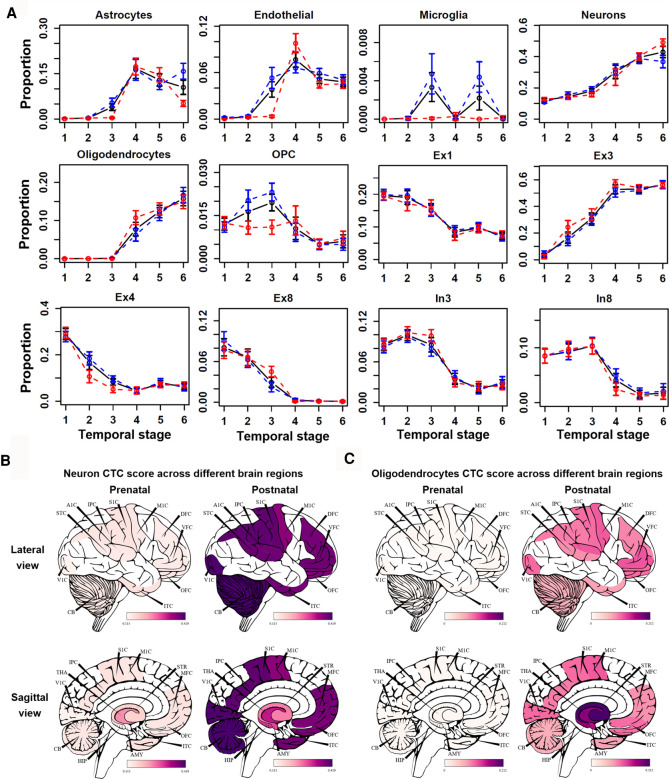
By using the CTC scores, we first studied the dynamic changes of CTC over time. By comparing the CTC scores with developmental stages, we found that the proportion of most fetal cell types (e.g., fetal astrocyte, fetal intermediate progenitor cell [IPC], fetal quiescence) decreased with age, whereas other cell types showed unique trends. As expected, fetal cell types were enriched at early fetal stages and were gradually replaced by nonfetal cell types along developmental stages (Supplemental Fig. S6). This result showed the validity of our analysis. We found that the proportion of neurons and oligodendrocytes was positively correlated with temporal stage (Fig. 5). OPC cells and oligodendrocytes showed inverse CTC score patterns. OPC cells, the progenitor cells of oligodendrocytes (Zhang et al. 2019b), showed high CTC scores at the prenatal stages (temporal stages 1–3), whereas oligodendrocytes showed high CTC scores at postnatal stages (temporal stages 4–6). Moreover, some cell types showed difference between males and females. For example, we observed a higher proportion of microglia cells at the intermediate postnatal stage (temporal stage 5) in males compared with females (Fig. 5A). This is consistent with a previous report (VanRyzin et al. 2020) that androgen-induced increase in endocannabinoid tone promoted microglia phagocytosis during a critical period of AMY development. Consistently, we found that the astrocyte composition in females was significantly higher than in males (median increase 0.04, t-test P = 0.04) at the intermediate postnatal stage (temporal stage 5). These findings suggested that sexual difference in CTC existed across the spatiotemporal context of normal human brain development. Our results thus provided insights into the underlying cellular mechanisms determining sexual difference in social behavior (McKay et al. 2014; VanRyzin et al. 2019), as well as etiology, epidemiology, and manifestation of psychiatric diseases (Aleman et al. 2003; Abel et al. 2010).
Figure 5.
Temporal and spatial changes in different cell-type compositions. (A) The black, blue, and red lines correspond to average CTC scores in all, male, and female individuals. Error bar, SD of replicates samples. Anatomical visualization of neuron (B) and oligodendrocyte (C) CTC score change between prenatal and postnatal groups across different regions.
We also examined the CTC changes across different brain regions. As shown in Figure 5B, the CTC scores of neurons increased in cerebral areas and the cerebellum but not in the diencephalon region. In contrast, the CTC scores of oligodendrocytes increased preferentially in the diencephalon region compared with the brain cortex region (Fig. 5C), which is consistent with the latest finding that oligodendrocytes are enriched in substantia nigra (Agarwal et al. 2020). In addition, the decrease of fetal quiescent CTC scores in the diencephalon region and cerebellum cortex was lower than in the brain cortex region; the CTC score of astrocytes in CBC only increased 0.02 compared with 0.13 (median) in the other 15 brain regions. Although age was primarily associated with gray matter thickness and fractional anisotropy of water diffusion in white matter tracts (McKay et al. 2014), our CTC spatial dynamic analysis implied that the development of the brain has strong spatial specificity. More results for spatial specificity of CTC in same group are presented in Supplemental Figure S7.
Neuronal subtype composition changes across temporal stage and region
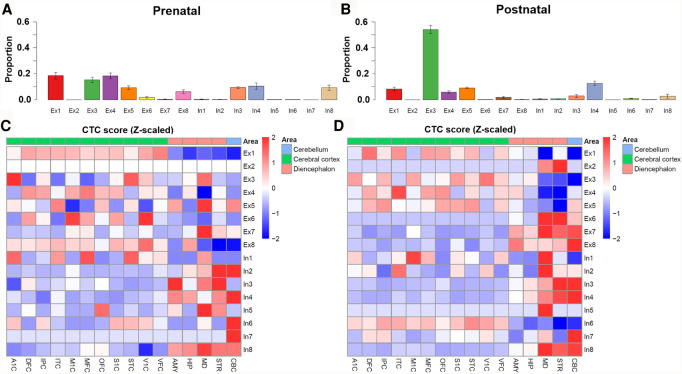
To further investigate how different neuronal cell types changed over brain development, we took advantage of single-cell reference panel B, which provided expression profiles for eight excitatory (Ex) neurons and eight inhibitory (In) neurons (Lake et al. 2016). Indeed, prominent differences were observed for the neuronal subtype composition between the prenatal and postnatal samples (Fig. 6A,B). For example, the proportion of Ex3 neuron, which was previously reported to be primarily enriched in the visual cortex (Lake et al. 2016), was significantly higher in postnatal individuals and may represent the increased importance of vision in postnatal compared with prenatal stages. In contrast, the proportions of Ex1, Ex4, and Ex8 negatively correlated with temporal stage (Fig. 5). In addition, some neuronal subtypes, for example, In3 and In8, peaked at the intermediate or late prenatal stage.
Figure 6.
Neuronal subtype deconvolution analysis. Relative proportion of neuronal subtype cell composition in prenatal (A) and postnatal (B). Error bar, SD of CTC scores across different patients and regions. Because the cell-type scores differed in several scale orders, we took z-score as the normalized CTC score to elucidate the correlation between neuronal subtype and brain region: (C) prenatal group and (D) postnatal group. We further separated 16 brain regions into three categories, (1) cerebral cortex (A1C, DFC, IPC, ITC, M1C, MFC, OFC, S1C, STC, V1C, VFC), (2) cerebellum (CBC), and (3) diencephalon region (AMY, HIP, MD, STR) for better visualizing.
We next investigated the neuronal subtypes in different brain regions. A direct comparison of each neuronal subtype proportion in each brain region revealed distinguishable preferences (Fig. 6C,D). Overall, most excitatory neuronal subtypes were enriched in the cerebral cortex, which is consistent with the recent finding that few excitatory neurons are enriched in the substantia nigra (Agarwal et al. 2020). For example, neuronal subtype Ex1 was enriched in the prefrontal cortex (DFC, MFC, OFC, and VFC) and temporal-parietal cortex regions (IPC, ITC, and STC), which was consistent with a previous study reporting that Ex1 was mainly enriched in brain regions BA21, BA22 (temporal cortex) and BA41/BA42 (frontal cortex) (Lake et al. 2016). In addition, Ex3 (specific to the visual cortex region) showed the highest enrichment in V1C in the postnatal group; Ex4 was enriched in all prefrontal cortex regions (DFC, MFC, OFC, and VFC) and had the highest proportion in ITC; and Ex5 was enriched in OFC and STC. There was also a difference in the proportion of neuronal subtypes between prenatal and postnatal groups. For example, the CTC score of Ex3 in the A1C region was higher than in the V1C region in the prenatal group, which implied dynamic changes during human brain development. On the other hand, we found most inhibitory neuronal subtypes were enriched in subcortical or cerebellum regions, for example, In3 in the STR region, In4 and In7 in the cerebellum, In5 in the MD, and In8 in the STR, CBC, and MD. These inhibitory neuronal subtype and brain region enrichment patterns were observed in both the prenatal (Fig. 6C) and postnatal (Fig. 6D) stages. These observations were consistent with previous reports that interneurons connecting the neocortex were largely inhibitory and were generated by progenitors in the subpallial (ventral) proliferative zone of the telencephalon before migrating into the neocortex (Cobos et al. 2001; Wichterle et al. 2001; Wonders and Anderson 2006). More results for spatial specificity of CTC in same group are presented in Supplemental Figure S8.
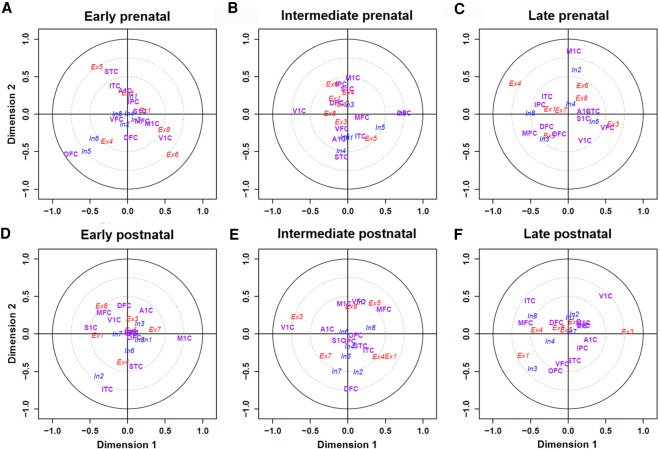
To further investigate neuronal-subtype-based spatiotemporal specificity on cerebral cortex areas, we applied canonical correspondence analysis (CCA). CCA infers information from two matrices and projects data points into a single embedding space. As shown in Figure 7, the distance from the center reflects the relationship strength, and the closeness of data points reflects the strong correlations. For better visualization, we used two-dimensional scatter plots, also known as canonical loading plots, to show the correspondence between cell types and brain regions (see Methods). Several tightly coupled relationships between neuronal subtypes and brain regions reinforced that different neuronal subtypes were likely involved in different functions. For example, neuronal subtype Ex3 is located close to the V1C region at the intermediate and late postnatal stages (Fig. 7E,F) but not at the prenatal and early postnatal stages (Fig. 7A–D), implying a stage-specific role of Ex3 cells in vision. We thus conducted functional enrichment analyses using GO and KEGG pathways for marker genes of each neuronal subtype. As presented in Supplemental Table S4, we found that Ex1-specific genes (n = 244) were mainly enriched in postsynaptic density (false-discovery rate [FDR] = 5 × 10−5), microtubule cytoskeleton organization and cytoplasm (FDR = 7 × 10−5), and regulation of synaptic plasticity pathways (FDR = 9 × 10−4). Ex3-specific genes (n = 206) were mainly enriched in cytosol (FDR = 6 × 10−5), protein binding (FDR = 2 × 10−3), and extracellular exosome pathways (FDR = 0.048). And In4-specific genes (n = 173) were mainly enriched in central nervous system development (FDR = 0.07).
Figure 7.
Visualization of the canonical correspondence analysis (CCA) results of neuronal subtypes across different brain regions under six growth stages. Assuming that both cell types and regions have unit variance, their projections on the plane reside within a circle of radius one centered at the origin. Distance to the center refers to the strength of the relationship. The first group (colored in red) consists of eight excitatory neuronal subtypes. The second group (blue) consists of eight inhibitory neuronal subtypes. The third group (magenta) consists of 11 brain regions on cerebral cortex hemispheres. For clarity, two circles with radii of 0.5 and 0.75 are shown to distinguish associations of cell types and regions. CCA results revealed a number of neuronal subtypes in close proximity to specific regions at six different temporal stages: (A–C) early, intermediate, and late prenatal stage; (D–F) early, intermediate, and post-postnatal stage.
Cell type–specific enrichment analysis of 13 major brain-associated traits
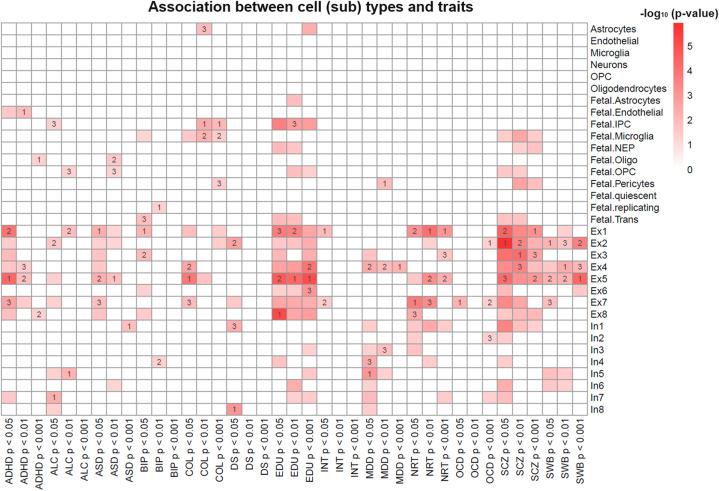
We collected GWAS summary statistics for 13 brain-associated traits and conducted cell type–specific enrichment analysis (Supplemental Table S5). These 13 traits included lots of psychiatric disorders, including attention deficit hyperactivity disorder, ASD, bipolar disorder, major depressive disorder, and SCZ. To extend our previous tissue-level study (Pei et al. 2019), we found that for most neuropsychiatric diseases, their susceptible genes were mainly enriched in excitatory and inhibitory neurons (Fig. 8). Specifically, we observed more excitatory neurons than inhibitory neurons enriched for genes associated with SCZ, education, neuroticism, ASD, and subjective wellbeing. These findings are consistent with recent study that reported the enrichment of excitatory over inhibitory neurons for SCZ (Finucane et al. 2018). On the other hand, we observed more inhibitory than excitatory neurons that were associated with major depressive disorder. Although previous study revealed same pattern for bipolar disorder (Finucane et al. 2018), the results varied slightly with different thresholds among trait-associated genes.
Figure 8.
Association between trait-associated genes and cell-type marker genes. The color scheme reflects the −log10 transformed P-value. The values 1, 2, and 3 in some cells indicated the rank of the corresponding cell type based on the Fisher's exact test. Nonsignificant associations (P-value ≥ 0.05) were replaced by blank color.
Our results from the cell type–specific enrichment analysis were consistent with previous reports (Parikshak et al. 2013; Willsey et al. 2013). These results may shed light onto the roles of neuronal subtypes in the manifestation of the GWAS signals. For example, SCZ-related genes were previously reported to be highly expressed during prenatal development (Gilman et al. 2012). In our results, we found that SCZ genes were enriched for the Ex1 (P = 3.2 × 10−5) and Ex4 (P = 5.5 × 10−4) neuronal subtypes, and these two neurons were also significantly enriched in the prenatal frontal cortex region (Fig. 6C). In addition, bipolar disorder–related genes were enriched in Ex1 (P = 5.7 × 10−3) and In4 (P = 0.04). Of note, neuron In4 was enriched for prenatal front amygdala and hippocampus regions (Fig. 6C), and the amygdala, hippocampus, and prefrontal cortex regions were previously reported to be related with bipolar disorder (Ellison-Wright and Bullmore 2010; Chai et al. 2011). In addition, ASD, number of education years, and college status were all enriched in fetal cell types (P < 0.05) (Fig. 8). Genes related to ASD and intellectual ability have long been implicated in early brain development (Shaw et al. 2006; Kelleher and Bear 2008; Coe et al. 2019), and our results further confirmed these conclusions at the cell-type level. Similarly, SCZ was also enriched in fetal cells. Collectively, these results revealed a cell-type-level enrichment of GWAS genes for several brain disorders, presenting a new way to interpret GWAS results in the context of spatiotemporal information from the human developing brain.
Discussion
scRNA-seq was shown as one of the most popular strategies to elucidate the heterogeneity of the human brain at the molecular level (Darmanis et al. 2015; Lake et al. 2016). However, human brain samples are extremely hard to collect, and there is a high level of incompleteness for human spatiotemporal transcriptome. In this study, we provided a reference grid of spatiotemporal transcriptome data in human brain using tensor-based imputation and BrainSpan data. We further gained a comprehensive understanding of human brain development and related brain disorders by conducting a series of genomics analyses. The currently available transcriptome data for human brain have high missing data rates. Our implemented tensor-based (Khan and Ammad-ud-din 2016) imputation could reliably infer a completed grid of spatiotemporal transcriptome data in BrainSpan with high accuracy (median: R2 > 0.9). The resulting imputed data provide a reliable resource for downstream analysis. Moreover, our application of deconvolution algorithms (Newman et al. 2015) to recent scRNA-seq data (Bakken et al. 2018; Wu et al. 2019) enabled us to systematically investigate brain CTC from both prenatal (range from 12 pcw to 38 pcw) and postnatal (4 mo to 40 yr) stages. The alterations in neuronal subtype composition across the entire range of human brain development yielded several interesting results that were beyond traditional bulk-tissue genomic analyses. We characterized brain CTC and discovered many neuronal subtypes showing temporal as well as spatial specificity. In contrast to previous studies, our results are derived from a large number of individuals, covering a diverse spectrum of temporal and spatial stages. This increased the generalizability of our findings.
To our best knowledge, this study provides the most comprehensive map of spatiotemporal CTC of the human brain. By integrating human brain spatiotemporal transcriptomics and brain disease–relevant GWAS data, we systematically interpreted the cell-type spatiotemporal specificity throughout human brain development and uncovered disease-relevant cell types for the major psychiatric disorders. Based on the neuronal subtype composition and functional enrichment of marker genes, we inferred the specific function of each neuronal subtype. For example, previous studies showed that the frontal cortex is mainly involved in concentration, planning, judgment, emotional expression, and creativity (Banks et al. 2007; de Souza et al. 2014); the primary visual cortex is mainly involved in vision; and the CBC is mainly involved in coordination of movement balance and equilibrium (Jacobs et al. 2018). However, with the exception of Ex3 being significantly enriched in the V1C region at the intermediate and late postnatal stages, the majority of neuronal subtypes was uniformly enriched for multiple brain functional areas. One possible reason is likely because most cellular signaling involves a diverse repertoire of cell types under precise molecular regulation (Miller et al. 2014). Moreover, neuronal subtype classification is mainly based on brain layer, as opposed to brain regions (Briggs 2010). A previous study revealed that the cerebral cortex is composed of six layers characterized by differences in the composition of various neuronal subtypes (Zeng et al. 2012). For example, layer 4 neurons are the main targets of thalamocortical inputs, whereas layer 5 and 6 neurons mainly transmit output projections to various subcortical and contralateral regions (Briggs 2010).
There are still potential limitations to improve upon. First, current deconvolution method depends on feature selection and the choice of reference set. Because the adult cells in scRNA-seq data are unlikely to appear in fetal tissues (and vice versa) and because there are very different cell populations between the substantia nigra and cortex region, using suitable reference scRNA-data for deconvolution analysis is very important (Sosina et al. 2020). However, owing to the minute amount of starting material, scRNA-seq data are prone to batch effects (Haghverdi et al. 2018). For better-selected cell-type signature genes, an integration of different large-scale scRNA-seq data generated by various groups in diverse experiments (Simon et al. 2020) would be an essential step for better deconvolution analysis. Second, compared with other imputation approaches (Huckins et al. 2019; Zhang et al. 2019a), our tensor-based model fail to consider genetic variation (Gusev et al. 2018), sexual differences (VanRyzin et al. 2019), and pregnancy events (Hoekzema et al. 2017). The imputation performance for a minor fraction of samples is still low. We speculate that an integrative model would further improve the performance. Nevertheless, we hope that our results will be useful for biomarker research in psychiatric disorders. Additionally, our analysis strategy may facilitate the generation of novel biological insights underlying brain diseases, and it can be applied to future data. Furthermore, novel technologies, such as RNA tomography (Junker et al. 2014; Wu et al. 2016), will generate more spatiotemporal gene expression features in tissues or organs. Our method can be further applied to such data, leading to a deeper understanding of the function of the human brain or other types of organs.
Methods
Neurodevelopmental transcriptome data
Bulk brain transcriptome data
The transcriptomic data (RNA-seq) of the developing human brain was downloaded from the Allen Institute BrainSpan Atlas (access date May 21, 2019). The raw data comprised 524 transcriptomes from 42 individuals ranging in age between 8 pcw and 40 yr across 26 brain regions. The overall missing data rate was ∼52% (568 out of 42 × 26 = 1092 transcriptomes), with 16/26 regions sampled for at least 20 individuals and 35/42 individuals sampled for at least five regions. After removing individuals with high missing data rates (i.e., ≥15%), we obtained a working data set with 35 individuals for 16 regions. Hereafter, we refer one sample as a transcriptome that was measured at a particular developmental stage in a particular region. One sample has one transcriptome, and an individual has multiple samples corresponding to multiple brain regions. The full names of each region as well as other details are presented in Supplemental Table S1. We also downloaded the transcriptome data from the GTEx consortium for adult human brain (downloaded on August 15, 2019).
Brain scRNA-seq data
scRNA-seq profiles were downloaded from PsychENCODE (Akbarian et al. 2015). Based on the original platforms, we defined two panels as the reference scRNA-seq data for brain. The first panel (denoted as panel A) was a combined data set using the scRNA-seq data derived from the cortical tissue of eight adults and four embryonic samples ranging from 16 to 18 gestational weeks (Darmanis et al. 2015) and PsychENCODE (Akbarian et al. 2015). The cell types in panel A included astrocytes, microglia, neurons, oligodendrocytes, oligodendrocyte precursor cells (OPC), and endothelial cells, as well as fetal astrocyte, endothelial, IPC, microglia, neuroepithelium (NEP), oligodendrocyte, fetal OPC, pericyte, quiescent, and replicating cells. The second panel (denoted as panel B) was derived from six cortical regions of a normal, 51-yr-old female postmortem brain by single-nucleus RNA sequencing (snRNA-seq) (Lake et al. 2016), including eight excitatory and eight inhibitory neuronal subtypes.
BrainSpan data imputation based on tensor decomposition
We transformed the BrainSpan transcriptome data with missing values into a 3D tensor , with axes corresponding to the individual, region, and protein-coding gene, respectively. Because all individuals have their corresponding ages, the axis for individuals also represented the dimension of lifespan. Thus, the BrainSpan data imputation problem was transformed as a tensor decomposition and completion task. We applied the Bayesian tensor decomposition method using the trilinear CP factorization (Khan and Ammad-ud-din 2016) algorithm. The CP method factorized an input tensor into a low-dimensional component space U, V, and W vector (rank-1 tensor), corresponding to the individual, region, and gene tensor, respectively. The number of components, denoted by R, was tested from 10 to 100. In our case, R was automatically determined and optimized by the package. The factorized tensors were optimized to approximate the measured data by minimizing reconstructed variance. The CP decomposition can be written as follows:
where R is a positive integer that represents the rank of the tensor, and ur, vr, wr denote rank-1 tensor with appropriate dimensions. Here, the notation “°” represents the outer product of tensors. The optimization function is defined as , where .
After model fitting, we used the factorized tensors to reconstruct the original tensor, which included not only the approximation of the measured samples but also the newly generated values for the originally missing samples. To obtain robust results, we repeated the imputation procedure 100 times and used the median values as final results for downstream analysis.
We performed LOO cross-validation to evaluate the results. For each sample with measured transcriptome data, we constructed a tensor excluding this sample (i.e., the holdout sample), applied CP factorization, and imputed the missing transcriptome. Imputation performance was evaluated using four measurements: PCC, R2, root mean squared error (RMSE), and mean absolute error (MAE).
Variance partition analysis by linear mixed model
After imputation, we organized the completed BrainSpan data as a matrix, with genes as rows (n = 18,921) and samples as columns (n = 560 for 35 individuals across 16 regions). Lowly expressed genes with RPKM ≤ 1 in >20% of the samples were excluded, resulting in 13,260 high-abundance genes for the following analyses. All gene expression values, including both measured and imputed, were z-score transformed. To assess the impact of several covariates, we applied a linear mixed model as implemented in the R package variancePartition (v1.8.1) (Hoffman and Schadt 2016), which decomposes the expression of each gene into components attributable to each variable. In this study, we considered the following biological or technical factors: individual, imputation status, sex, and spatial and temporal information. Categorical variables were modeled as random effects. For temporal information, we grouped all samples into six stages, corresponding to early prenatal (before 12 pcw), intermediate prenatal (from 13–21 pcw), late prenatal (after 24 pcw), early postnatal (4 mo–4 yr), intermediate postnatal (8–13 yr), and late postnatal (18–40 yr) stages. A model was fitted for each gene independently, and the results for all genes were aggregated afterward. The results were visualized using the built-in function plotPercentBars of the variancePartition package (Hoffman and Schadt 2016).
WGCNA
WGCNA (v1.67) (Langfelder and Horvath 2008) was used to build a coexpression network based on the imputed BrainSpan RNA-seq profiles. The modules with a PCC > 0.5 and a correlation test P-value < 1 × 10−10 were extracted as temporal or spatial associated modules.
Gene set enrichment analysis
We used RDAVIDWebService (v 1.19.0) (Fresno and Fernandez 2013) for gene set enrichment analysis. All human protein-coding genes were used as the background gene set. Benjamini and Hochberg's approach (Benjamini and Hochberg 1995) was used for multiple test correction. Significant pathways were defined as those with adjusted P-value < 0.001. We also conducted TSEA using the deTS method (Pei et al. 2019; Jia et al. 2020) and the GTEx panel as the reference, without multiple test correction.
Cell composition deconvolution
For bulk transcriptome data, including both measured and imputed RNA-seq expression data, we used CIBERSORT (v1.04) (Newman et al. 2015) relative mode to perform deconvolution and quantify CTCs. CIBERSORT uses a reference single-cell expression panel and implements a support vector regression (SVR)–based machine learning approach to estimate the composition of each cell type. Here, we applied CIBERSORT to the BrainSpan data set with the two single-cell data sets aforementioned. To balance the number of cells, we randomly selected 50 cells in each cell type. For a few cell types with fewer than 50 cells, all cells were used. To balance the batch effect between two panels (Supplemental Fig. S9), we deconvoluted them using the separate reference panel. Panel A had 715 signature genes. Panel B had 623 signature genes (Supplemental Table S6). The bulk transcriptome and the expression signatures of cell types were simultaneously submitted to the CIBERSORT pipeline. For each bulk transcriptome, we obtained a CTC score for each cell type from the reference panel. We used the R package cerebroViz (Bahl et al. 2017) for anatomical visualization of spatiotemporal brain data. We applied PCA to visualize the distribution of CTC scores.
CCA
CCA is a multivariate technique to identify the relationship between two sets of explanatory variables (Hotelling 1936). Specifically, CCA projects the two variables onto a low-dimensional space where these variables are maximally correlated. In our case, we used CCA to investigate the relationships between the variable of brain regions and the variable of cell types. Let X be an N × C matrix of CTC scores in N samples (i.e., transcriptomes, in which a transcriptome was obtained from a sample in a particular brain region at a particular developmental stage) and C cell types. Similarly, let Y denote an N × R matrix recording the source of each transcriptome in R brain regions, where yn,r = 100% (i.e., the value one) indicates the nth (n = 1,…,N) transcriptome was collected from the rth (r = 1,…,R) region. Let and denote the two basis vectors. Then the projections of the two explanatory variables onto these basis vectors are given by
and
CCA seeks to find two vectors (a and b) to maximize the correlation . Thus, the correlations between two projections are mutually maximized as follows:
where the derived linear projections U1 and V1 are the first canonical components, and ρ1 refers to the canonical correlation between the first components. Note that the successively computed canonical correlations decrease by nature; that is, ρ1 ≥ ρ2 ≥ … ≥ ρmin(C,R). The CCA results presented in this work is conducted by the R package CCA (Gonzalez et al. 2008).
Trait-associated genes from GWAS
We downloaded GWAS data sets for 13 brain-associated traits (for details, see Supplemental Table S6). To define trait-associated genes, we mapped SNPs to genes if they are located in the gene body or 50 kb upstream of or downstream from the gene using the PASCAL software (Lamparter et al. 2016). PASCAL calculated a gene-based P-value while taking into account linkage disequilibrium, gene length, and SNP density. For each trait, we defined trait-associated genes based on the gene-based P-values using three thresholds to ensure rigor: P-value < 0.05, 0.01, and 0.001.
Cell type–specific enrichment analysis
To conduct cell type–specific enrichment analysis for a list of disease genes, we merged the single-cell expression profiles from both panels of cell-type data. Briefly, we fitted a regression model for each gene to assess its expression specificity in each cell type using: Y ∼ X, where Y = [yi], i = 1,…, N was a vector of the normalized gene expression in a total of N cells, and X was the cell-type group status. Specifically, for a cell type in examination, we defined X = [xi], i = 1,…, N, where xi = 1 if the sample belongs to the cell type in examination and xi = 0 otherwise. After model fitting, we obtained t-statistics that can be used to measure the cell-type specificity for each gene. A high t-statistic value indicates that the gene is specifically expressed in the corresponding cell type. Based on our previous work (Pei et al. 2019), we defined the top 5% of genes ordered by decreasing t-statistic as the cell type–specific genes. Fisher's exact test was then used to evaluate the association between trait-associated genes and cell type–specific genes.
Software availability
Source code implementing all steps, including data preprocessing, tensor imputation, evaluation scripts, and the completed BrainSpan data set, is available via GitHub (https://github.com/bsml320/BrainSpan) and as Supplemental Code.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Ackowledgments
We thank the members of Bioinformatics and Systems Medicine Laboratory (BSML) for valuable discussion. This work was partially supported by National Institutes of Health grants (R01LM012806, R03DE027393, R03DE028103, and R03DE027711) and the Cancer Prevention and Research Institute of Texas grant (CPRIT RP180734). Publication charges for this article have been funded by R01LM012806.
Author contributions: G.P., P.J., and Y.-Y.W. conceived the study. G.P. performed all bioinformatics analysis. L.M.S. helped with the results discussion and English editing. Y.D. helped with GWAS data collection. G.P., P.J., and Z.Z. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.265769.120.
References
- Abel KM, Drake R, Goldstein JM. 2010. Sex differences in schizophrenia. Int Rev Psychiatry 22: 417–428. 10.3109/09540261.2010.515205 [DOI] [PubMed] [Google Scholar]
- Agarwal D, Sandor C, Volpato V, Caffrey TM, Monzón-Sandoval J, Bowden R, Alegre-Abarrategui J, Wade-Martins R, Webber C. 2020. A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat Commun 11: 4183 10.1038/s41467-020-17876-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, et al. 2015. The PsychENCODE project. Nat Neurosci 18: 1707–1712. 10.1038/nn.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman A, Kahn RS, Selten JP. 2003. Sex differences in the risk of schizophrenia: evidence from meta-analysis. Arch Gen Psychiatry 60: 565–571. 10.1001/archpsyc.60.6.565 [DOI] [PubMed] [Google Scholar]
- Aran D, Hu Z, Butte AJ. 2017. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18: 220 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl E, Koomar T, Michaelson JJ. 2017. cerebroViz: an R package for anatomical visualization of spatiotemporal brain data. Bioinformatics 33: 762–763. 10.1093/bioinformatics/btw726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Hodge RD, Miller JA, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, et al. 2018. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One 13: e0209648 10.1371/journal.pone.0209648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks SJ, Eddy KT, Angstadt M, Nathan PJ, Phan KL. 2007. Amygdala-frontal connectivity during emotion regulation. Soc Cogn Affect Neurosci 2: 303–312. 10.1093/scan/nsm029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Gazzaniga MS. 2011. Understanding complexity in the human brain. Trends Cogn Sci 15: 200–209. 10.1016/j.tics.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Briggs F. 2010. Organizing principles of cortical layer 6. Front Neural Circuits 4: 3 10.3389/neuro.04.003.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai XJ, Whitfield-Gabrieli S, Shinn AK, Gabrieli JD, Nieto Castañón A, McCarthy JM, Cohen BM, Öngür D. 2011. Abnormal medial prefrontal cortex resting-state connectivity in bipolar disorder and schizophrenia. Neuropsychopharmacology 36: 2009–2017. 10.1038/npp.2011.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Puelles L, Martínez S. 2001. The avian telencephalic subpallium originates inhibitory neurons that invade tangentially the pallium (dorsal ventricular ridge and cortical areas). Dev Biol 239: 30–45. 10.1006/dbio.2001.0422 [DOI] [PubMed] [Google Scholar]
- Coe BP, Stessman HAF, Sulovari A, Geisheker MR, Bakken TE, Lake AM, Dougherty JD, Lein ES, Hormozdiari F, Bernier RA, et al. 2019. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet 51: 106–116. 10.1038/s41588-018-0288-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, et al. 2011. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478: 519–523. 10.1038/nature10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, Morgan J. 2007. Mapping early brain development in autism. Neuron 56: 399–413. 10.1016/j.neuron.2007.10.016 [DOI] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. 2015. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci 112: 7285–7290. 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza LC, Guimarães HC, Teixeira AL, Caramelli P, Levy R, Dubois B, Volle E. 2014. Frontal lobe neurology and the creative mind. Front Psychol 5: 761 10.3389/fpsyg.2014.00761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haene E, Jacobs EZ, Volders PJ, De Meyer T, Menten B, Vergult S. 2016. Identification of long non-coding RNAs involved in neuronal development and intellectual disability. Sci Rep 6: 28396 10.1038/srep28396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham TJ, Libbrecht MW, Howbert JJ, Bilmes J, Noble WS. 2018. PREDICTD PaRallel epigenomics data imputation with cloud-based tensor decomposition. Nat Commun 9: 1402 10.1038/s41467-018-03635-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison-Wright I, Bullmore E. 2010. Anatomy of bipolar disorder and schizophrenia: a meta-analysis. Schizophr Res 117: 1–12. 10.1016/j.schres.2009.12.022 [DOI] [PubMed] [Google Scholar]
- Farahany NA, Greely HT, Hyman S, Koch C, Grady C, Pașca SP, Sestan N, Arlotta P, Bernat JL, Ting J, et al. 2018. The ethics of experimenting with human brain tissue. Nature 556: 429–432. 10.1038/d41586-018-04813-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane H, Reshef Y, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh P-R, Lareau C, Shoresh N, et al. 2018. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50: 621–629. 10.1038/s41588-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno C, Fernandez EA. 2013. RDAVIDWebService: a versatile R interface to DAVID. Bioinformatics 29: 2810–2811. 10.1093/bioinformatics/btt487 [DOI] [PubMed] [Google Scholar]
- Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. 2012. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci 15: 1723–1728. 10.1038/nn.3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glastonbury CA, Couto Alves A, El-Sayed Moustafa JS, Small KS. 2019. Cell-type heterogeneity in adipose tissue Is associated with complex traits and reveals disease-relevant cell-specific eQTLs. Am J Hum Genet 104: 1013–1024. 10.1016/j.ajhg.2019.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Fernandez O, Fernandez R, Menendez C, Maza J, Gonzalez-Quevedo A, Buergo MA. 2008. Association between blood lipids and types of stroke. MEDICC Rev 10: 27–32. 10.37757/MR2008.V10.N2.9 [DOI] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, Song L, Safi A, McCarroll S, Neale BM, et al. 2018. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet 50: 538–548. 10.1038/s41588-018-0092-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, Marioni JC. 2018. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36: 421–427. 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekzema E, Barba-Müller E, Pozzobon C, Picado M, Lucco F, García-García D, Soliva JC, Tobeña A, Desco M, Crone EA, et al. 2017. Pregnancy leads to long-lasting changes in human brain structure. Nat Neurosci 20: 287–296. 10.1038/nn.4458 [DOI] [PubMed] [Google Scholar]
- Hoffman GE, Schadt EE. 2016. Variancepartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17: 483 10.1186/s12859-016-1323-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. 1936. Relations between two sets of variates. Biometrika 28: 321–377. 10.1093/biomet/28.3-4.321 [DOI] [Google Scholar]
- Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardinas AF, Rajagopal VM, Als TD, Nguyen HT, Girdhar K, et al. 2019. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet 51: 659–674. 10.1038/s41588-019-0364-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HIL, Hopkins DA, Mayrhofer HC, Bruner E, van Leeuwen FW, Raaijmakers W, Schmahmann JD. 2018. The cerebellum in Alzheimer's disease: evaluating its role in cognitive decline. Brain 141: 37–47. 10.1093/brain/awx194 [DOI] [PubMed] [Google Scholar]
- Jia P, Han G, Zhao J, Lu P, Zhao Z. 2017. SZGR 2.0: a one-stop shop of schizophrenia candidate genes. Nucleic Acids Res 45: D915–D924. 10.1093/nar/gkw902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Chen X, Fanous AH, Zhao Z. 2018. Convergent roles of de novo mutations and common variants in schizophrenia in tissue-specific and spatiotemporal co-expression network. Transl Psychiatry 8: 105 10.1038/s41398-018-0154-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Dai Y, Hu R, Pei G, Manuel AM, Zhao Z. 2020. TSEA-DB: a trait–tissue association map for human complex traits and diseases. Nucleic Acids Res 48: D1022–D1030. 10.1093/nar/gkz957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker JP, Noël ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J, van Oudenaarden A. 2014. Genome-wide RNA tomography in the zebrafish embryo. Cell 159: 662–675. 10.1016/j.cell.2014.09.038 [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature 478: 483–489. 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ 3rd, Bear MF. 2008. The autistic neuron: troubled translation? Cell 135: 401–406. 10.1016/j.cell.2008.10.017 [DOI] [PubMed] [Google Scholar]
- Khan SA, Ammad-ud-din M. 2016. tensorBF: an R package for Bayesian tensor factorization. bioRxiv 10.1101/097048 [DOI]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. 2016. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352: 1586–1590. 10.1126/science.aaf1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. 2016. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput Biol 12: e1004714 10.1371/journal.pcbi.1004714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 1471–2105. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DR, Knowles EE, Winkler AA, Sprooten E, Kochunov P, Olvera RL, Curran JE, Kent JW, Carless MA, Göring HH, et al. 2014. Influence of age, sex and genetic factors on the human brain. Brain Imaging Behav 8: 143–152. 10.1007/s11682-013-9277-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. 2014. Transcriptional landscape of the prenatal human brain. Nature 508: 199–206. 10.1038/nature13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. 2015. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12: 453–457. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. 2013. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155: 1008–1021. 10.1016/j.cell.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnavelas JG. 2000. The origin and migration of cortical neurones: new vistas. Trends Neurosci 23: 126–131. 10.1016/S0166-2236(00)01553-8 [DOI] [PubMed] [Google Scholar]
- Pei G, Dai Y, Zhao Z, Jia P. 2019. deTS: tissue-specific enrichment analysis to decode tissue specificity. Bioinformatics 35: 3842–3845. 10.1093/bioinformatics/btz138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta V, Cuesta MJ. 2017. Motor abnormalities: from neurodevelopmental to neurodegenerative through “functional” (neuro)psychiatric disorders. Schizophr Bull 43: 956–971. 10.1093/schbul/sbx089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, Evans A, Rapoport J, Giedd J. 2006. Intellectual ability and cortical development in children and adolescents. Nature 440: 676–679. 10.1038/nature04513 [DOI] [PubMed] [Google Scholar]
- Simon LM, Wang Y-Y, Zhao Z. 2020. INSCT: integrating millions of single cells using batch-aware triplet neural networks. bioRxiv 10.1101/2020.05.16.100024 [DOI]
- Sosina OA, Tran MN, Maynard KR, Tao R, Taub MA, Martinowich K, Semick SA, Quach BC, Weinberger DR, Hyde TM, et al. 2020. Strategies for cellular deconvolution in human brain RNA sequencing data. bioRxiv 10.1101/2020.01.19.910976 [DOI]
- VanRyzin JW, Marquardt AE, Argue KJ, Vecchiarelli HA, Ashton SE, Arambula SE, Hill MN, McCarthy MM. 2019. Microglial phagocytosis of newborn cells is induced by endocannabinoids and sculpts sex differences in juvenile Rat social play. Neuron 102: 435–449.e6. 10.1016/j.neuron.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRyzin JW, Marquardt AE, Pickett LA, McCarthy MM. 2020. Microglia and sexual differentiation of the developing brain: a focus on extrinsic factors. Glia 68: 1100–1113. 10.1002/glia.23740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. 2011. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474: 380–384. 10.1038/nature10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gamazon ER, Pierce BL, Stranger BE, Im HK, Gibbons RD, Cox NJ, Nicolae DL, Chen LS. 2016. Imputing gene expression in uncollected tissues within and beyond GTEx. Am J Hum Genet 98: 697–708. 10.1016/j.ajhg.2016.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Feng H, Ustianenko D, Duffié R, Yan Q, Jacko M, Martinez JC, Goodwin M, Zhang X, Hengst U, et al. 2018. Precise temporal regulation of alternative splicing during neural development. Nat Commun 9: 2189 10.1038/s41467-018-04559-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. 2001. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development 128: 3759–3771. [DOI] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. 2013. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155: 997–1007. 10.1016/j.cell.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. 2006. The origin and specification of cortical interneurons. Nat Rev Neurosci 7: 687–696. 10.1038/nrn1954 [DOI] [PubMed] [Google Scholar]
- Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, Noël ES, Grün D, Berezikov E, Engel FB, et al. 2016. Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev Cell 36: 36–49. 10.1016/j.devcel.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Wu H, Kirita Y, Donnelly EL, Humphreys BD. 2019. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 30: 23–32. 10.1681/ASN.2018090912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Liu X, Leng F, Li W. 2020. Blood-based multi-tissue gene expression inference with Bayesian ridge regression. Bioinformatics 36: 3788–3794. 10.1093/bioinformatics/btaa239 [DOI] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, et al. 2012. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 149: 483–496. 10.1016/j.cell.2012.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Voloudakis G, Rajagopal VM, Readhead B, Dudley JT, Schadt EE, Björkegren JLM, Kim Y, Fullard JF, Hoffman GE, et al. 2019a. Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits. Nat Commun 10: 3834 10.1038/s41467-019-11874-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu XY, Casella G, Tian J, Ye ZQ, Yang T, Han JJ, Jia LY, Rostami A, Li X. 2019b. Generation of oligodendrocyte progenitor cells from mouse bone marrow cells. Front Cell Neurosci 13: 247 10.3389/fncel.2019.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.