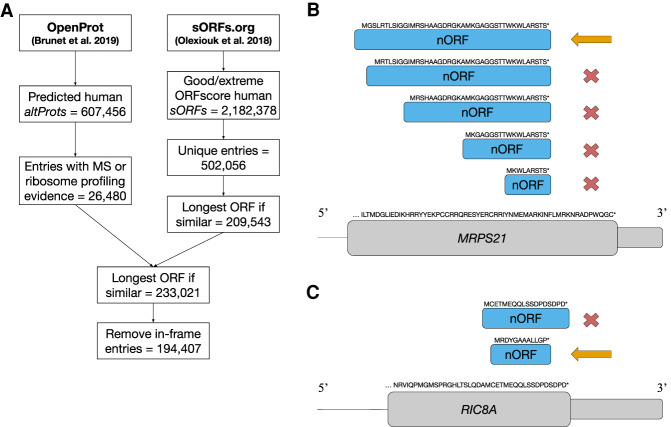
Figure 1.
Flow chart for the curation of the nORFs data set. (A) Steps illustrating the workflow to curate nORFs entries. From OpenProt, all predicted human altProts were filtered to entries with MS or ribosome profiling evidence (26,480). From sORFs.org, all human sORFs with an ORFscore of good or extreme were filtered to unique entries (502,056) and then summarized to the longest ORF at sites with multiple ORFs. Entries were then merged, the longest ORF was selected at multiple ORF sites, and in-frame entries were removed, leaving a total of 194,407 nORFs in the final data set. (B) An example of selecting the longest ORF for five small ORFs (smORFs) in an alternative frame of the final coding exon of the MRPS21 gene. In cases in which the ORFs share the same end site and differ only by their start site, we retain the longest ORF, indicated by the orange arrow, and remove the shorter ORFs, indicated by the red cross. (C) An example of removing in-frame entries in which two smORFs overlap the CDS of the RIC8A gene. The ORF in the same frame as the RIC8A CDS is removed from the data set as indicated by the red cross, whereas the second ORF in a different frame is retained in the data set, indicated by the orange arrow.