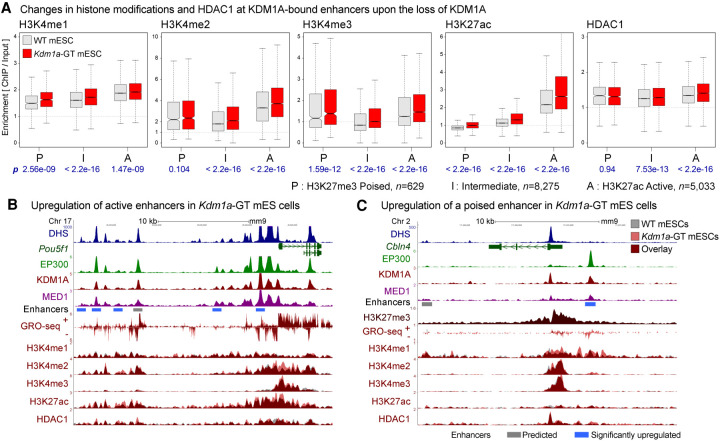
Figure 2.
Loss of KDM1A results in increases in H3K4 methylation and H3K27 acetylation at enhancers. (A) H3K4me1, H3K4me2, H3K4me3, H3K27ac, and HDAC1 levels on KDM1A-bound enhancers in WT (gray boxes) and Kdm1a-GT mESCs (red boxes). Enhancers were classified into poised (P), intermediate (I), and active (A) enhancers based on the enrichment of either H3K27ac or H3K27me3. Geometric mean of ChIP:Input ratios from the two independent ChIP-seq replicates are shown. P-values (p) from Wilcoxon signed-rank tests on log2(Kdm1a-GT/WT) are denoted in blue beneath each panel. n indicates the number of enhancers in each category. (B) Dysregulation of active enhancers at the Pou5f1 locus. Several enhancers are co-occupied by EP300 and KDM1A in WT mESCs, some of which show increased H3K4me2, H3K27ac, and GRO-seq signals in Kdm1a-GT mESCs (red vs. gray). (C) Misregulation of a poised enhancer at the Cbln4 locus. This locus is decorated with a broad H3K27me3 domain and shows elevations in H3K4me1, H3K4me2, and GRO-seq signals upon the loss of KDM1A. Gray bar: Predicted enhancer, blue bar: significantly up-regulated enhancer in Kdm1a-GT mESCs compared with WT mESCs based on changes in the GRO-seq signal (see Fig. 3).