Figure 1.
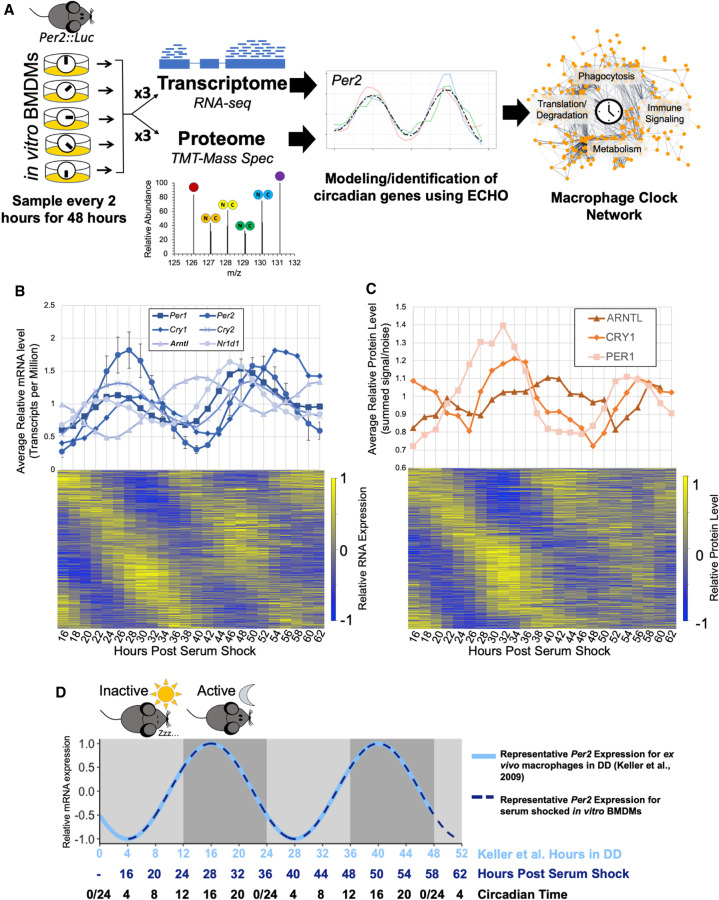
Multi-omics profiling details extensive circadian regulation of the macrophage transcriptome and proteome. (A) A schematic of the analysis of macrophage circadian regulation. Bone marrow derived macrophages (BMDMs) from Per2::Luc mice were synchronized in vitro via serum shock and sampled in triplicate for global proteome and transcriptome profiling with 10-plex tandem mass tag mass spectrometry and RNA-seq, respectively. Circadianly oscillating genes were identified using the ECHO program to determine the macrophage circadian protein–protein interaction network. (B) All detected clock gene transcripts oscillated circadianly, and error bars show the standard deviation for Per2 as a representative gene for the variation between triplicate values. Heat map in bottom panel shows relative expression for all identified circadian transcripts. (C) All detected clock proteins exhibited circadian abundances with delay from their respective transcripts. Points shown are an average of the summed signal/noise of peptides for the identified protein for up to three replicates depending on the detection at the given time point. Heat map in bottom panel shows relative expression for all identified circadian proteins. (D) A schematic summarizing how circadian time (CT) for our in vitro synchronized experimentation was inferred from comparison of Per2 mRNA oscillation timing we observed to the oscillation of Per2 mRNA reported in macrophages extracted from light-entrained mice transferred to constant dark conditions (Keller et al. 2009). Shading represents the relative inactive period (light gray) and active periods (dark gray) during the Dark:Dark (DD) experiment by Keller et al. Because mice are nocturnal, CT0/24 is the onset of the inactive period and CT12 is the onset of the active period.