ABSTRACT
Accumulating data manifest that long non-coding RNA (lncRNAs) are involved in all kinds of neurodegenerative disorders, consisting of the onset and progression of Alzheimer’s disease (AD). The study was for the research of the mechanism of lncRNA H19 (H19) in viability and apoptosis of PC12 cells induced by Aβ25-35 in a cellular model of AD with the regulation of microRNA (miR)-129 and high mobility group box-1 protein (HMGB1). An AD cellular model of PC12 cells was established using Aβ25-35. The Aβ25-35-induced PC12 cells were transfected with si-H19 or miR-129 mimic to figure their roles in cell viability,apoptosis, mitochondrial membrane potential dysfunction and oxidative stress in AD. Luciferase reporter assay and RNA-pull down assay were employed for verification of the binding relationship between H19 and miR-129 and the targeting relationship between miR-129 and HMGB1. An AD mouse model was induced and brain tissues were collected. H19, miR-129 and HMGB1 were detected in Aβ25-35-treated cells and brain tissues of AD mice. Elevated H19, HMGB1 and decreased miR-129 were found in Aβ25-35-treated PC12 cells as well as in brain tissues of AD mice. Silenced H19 or elevated miR-129 promoted viability, inhibited apoptosis, prevented mitochondrial membrane potential dysfunction and decreased oxidative stress in Aβ25-35-treated PC12 cells. H19 could specifically bind to miR-129. MiR-129 specifically suppressed HMGB1 expression. This study suggests that silenced H19 and up-regulated miR-129 accelerates viability and represses apoptosis of PC12 cells stimulated by Aβ25-35 in AD, which is beneficial for AD treatment.
KEYWORDS: Alzheimer’s disease, long non-coding RNA H19, MicroRNA-129, high mobility group box-1 protein, Aβ25-35, PC12 cells, apoptosis, viability
Introduction
Alzheimer’s disease (AD) is a long-term neurodegenerative disorder, the most prevalent reason of dementia in elderly people manifesting as dependence, disability, and mortality in patients [1]. The features of AD include a group of irreversible cognitive dysfunctions and neuropsychiatric symptoms [2]. The occurrence of AD is elevated gradually [3], and AD is influenced by plenty of elements, consisting of environmental factors and genetic and epigenetic variations [4]. Dementia in AD results in numerous reduction in production and living of the affected population [5]. AD is incurable and the drugs used for AD are only capable of relieving symptoms [6]. Therefore, in order to establish a new therapeutic method for AD, it is essential to explore the molecular mechanisms underlying the formation and progression of AD.
Long non-coding RNA (lncRNA) H19 is a preserved lncRNA that is situated in both the nucleus and the cytoplasm [7]. A study has found that elevated H19 in Schwann cells is implicated with neuropathic pain in the peripheral neuropathic pain [8]. In addition, a study has also revealed a new H19-modulated mechanism in seizure-stimulated neural damage and offers a novel target in developing lncRNA-based approaches to decrease seizure-induced brain injury in temporal lobe epilepsy (TLE) [9]. Moreover, an analysis review concludes that H19 delays neurogenesis after ischemic stroke [10]. MicroRNAs (miRNAs) are a classification of non-coding RNAs that are 20–24 nucleotides in length, with its biological functions by depressing the expression of target genes through base-pairing with endogenous mRNAs [11]. MiR-129 is an endothelial-specific miRNA that is significant in vascular integrity and angiogenesis [12]. miR-129-5p has been implicated as a possible choice for the treatment of AD [13], with regulatory effects on cognitive function and inflammation [14]. Significantly, modifying miR-129 could attenuate neuronal apoptosis and cognitive dysfunction in AD [15]. A study has indicated that miR-129-2 is able to directly target and depress high-mobility group box 1 (HMGB1) [16]. The pro-inflammatory cytokine HMGB1 is a multi-functional redox‐sensitive protein expressed and produced by all kinds of cells including bone cells, which manifests various cellular compartment‐specific functions [17]. A prior study has suggested that the therapeutic influences of the anti-HMGB1 monoclonal antibody are modulated by Aβ-independent mechanisms in AD [18]. Together, the data have indicated that depression of HMGB1 may be a possible therapeutic method for treating AD [19]. Therefore, the study was to explore the mechanism of H19 in proliferation and apoptosis of PC12 cells stimulated by Aβ25-35 in a cellular model of AD with the involvement of miR-129 and HMGB1.
Materials and methods
Ethics statement
This study was implemented with the Ethics Committee of College of Nursing, Inner Mongolia University for Nationalities and animals were treated humanly to minimize the sufferings.
AD cell modeling
PC12 cells (Cell resource center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China) were cultured in 10% fetal bovine serum (FBS)-Dulbecco’s Modified Eagle’s Medium (DMEM) (penicillin 100 kU/L, streptomycin 100 μg/mL). Cells maintaining 80–90% confluence were detached and passaged. Amyloid β-Protein Fragment 25–35 (Aβ25-35) was purchased from Sigma-Aldrich Chemical Company (St Louis, MO Missouri, USA). Aβ25-35 (1 mg) was dissolved in distilled water (1.8 mL) to reach 500 μmol/L and aggregated. The AD cell model was induced by Aβ25-35 [20,21]. PC12 cells in logarithmic growth state were starved with 1% serum DMEM 1 d before the experiment and stimulated by 25 μmol/L Aβ25-35 for 24 h [22]. The success of modeling was judged through observing cell morphology and growth state.
Observation of cell morphology
PC12 cells were trypsinized and seeded into 6-well plates at (1 ~ 1.5) × 102 cells/mL (each well containing 4 mL cell suspension). Serum-free treatment was performed 1 d before the experiment., Cells in the experimental group were joined with Aβ25-35 at 0 μmol/L, 12.5 μmol/L, 25 μmol/L and 50 μmol/L, respectively, while those in the control group with the same amount of culture medium. After 24 h, cell morphology was observed by a microscope.
Cell grouping and transfection
The cells were classified into the blank (only treated with Aβ25-35), the si-negative control (NC) (Aβ25-35 treatment and transfection with lncRNA-H19 siRNA NC), the si-H19 (Aβ25-35 treatment and transfection with lncRNA-H19 siRNA), the mimic NC (Aβ25-35 treatment and transfection with miR-129 mimic NC), the miR-129 mimic (Aβ25-35 treatment and transfection with miR-129 mimic), the overexpressed (OE)-H19 + mimic NC (Aβ25-35 treatment and transfection with up-regulated H19 vector + miR-129 mimic NC); the OE-H19 + miR-129 mimic (Aβ25-35 treatment and transfection with up-regulated H19 vector + miR-129 mimic) groups. All si-NC, si-H19, mimic NC, miR-129 mimic, OE-H19, and OE-NC were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Transfection was performed in line with the instructions of the LipofectamineTM 2000 (Invitrogen, CA, USA).
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
PC12 cells were detached and seeded into 96-well plates. Each well was supplemented with 200 μL cell suspension, with 3 duplicated wells per group. The culture medium (100 μL) was regarded as a blank control. Cells were reacted with MTT solution (20 μL/well) for 3 h and added with 150 μL Dimethyl Sulfoxide. Then, the optical density (OD) value was measured via the micro-plate reader. The cell viability = OD value of the test group/OD value of the blank group × 100%.
Colony formation assay
PC12 cells were detached and cultivated in 6-well plates at 400 cells/well for 2 wk to generate colonies. The cells were fixed, stained with 0.2% crystal violet solution and scanned.
Flow cytometry
Cells were fixed with 70% cold absolute ethanol and incubated with propidium iodide (PI, Sigma-Aldrich) with RNase A and evaluated for cell cycle distribution [23,24] by flow cytometry with cell quest software (FACS Calibur, BD Biosciences, USA).
Cells were detached with 0.25% trypsin (without ethylene diamine tetraacetic acid) (yb15050057, Yu Bo Biotech Co., Ltd., Shanghai, China) and centrifuged. Annexin-V-fluorescein isothiocyanate (FITC), PI and HEPES buffer solution were mixed to prepare Annexin-V-FITC/PI staining solution at 1:2:50 in the Annexin-V-FITC cell apoptosis detection kit (k201-100, Biovision, USA). Cells (1 × 106 cells) were suspended in 100 μL staining solution and mixed with 1 mL of HEPES buffer solution (pb180325, Procell, Wuhan, China). The fluorescence of FITC and PI was detected by excitation of 525 nm and 620 nm band-pass filter at 488 nm [25].
Hoechst 33258 staining
The cells were fixed and conducted with permeabilization with Triton-X100. Then, the cells were joined with 30 μL Hoechst 33258 solution (0.5 mol/mL) and sealed with the anti-fluorescence quenching agent. Cell images were captured under an inverted fluorescent biological microscope.
Intracellular mitochondrial membrane potential (MMP) measurement
The cells in 6-well plates at 1 × 106 cells/well were grown to 80% confluence and detached with trypsin. Then, cells were centrifuged at 1000 r/min, re-suspended in 0.5 mL culture medium and reacted with JC-1 staining solution. Finally, cells were subjected to centrifugation at 1000 r/min and immersion with JC-1 staining buffer (1 ×). The fluorescence intensity was tested via a flow cytometer.
Quantitative detection of intracellular reactive oxygen species (ROS)
The cells in 6-well plates at 1 × 106 cells/well were cultivated to 80% confluence, detached, centrifuged at 1000 r/min and incubated with 1 mL diluted 2,7-Dichlorodi-hydrofluorescein diacetate. The fluorescence intensity was measured via a flow cytometer.
Catalase (CAT), superoxide dismutase (SOD), and malondialdehyde (MDA) levels detection
CAT, SOD and MDA in cells were detected by CAT, SOD and MDA activity detection kits (JianCheng Bioengineering Institute, Nanjing, China (batch numbers were 20,141,120, 20,150,421, 20,150,223)). The cells (1 × 106) were seeded into 6-well plates per well. Cells maintaining 80% confluence were disrupted by ultrasound and centrifuged to collect the supernatant (200 μL). The OD value was measured at 240, 550, and 532 nm.
Fluorescence in situ hybridization (FISH)
Bioinformatics tools (http://lncatlas.crg.eu/) firstly analyzed the subcellular localization of H19. Cells were fixed with 4% paraformaldehyde, lysed with 0.5% Triton X-100 and hybridized with H19 probe. After that, cells were stained with 4’,6-diamidino-2-phenylindole 2hci and scanned with a fluorescence microscope (Olympus, Tokyo, Japan).
Dual-luciferase reporter gene assay
The binding sites of H19 and miR-129 and that of miR-129 and HMGB1 were predicted using the bioinformatics websites https://cm.jefferson.edu/rna22/Precomputed/and http://www.targetscan.org, respectively. The amplified wild-type (WT) of 3’-UTR H19 and 3’-UTR HMGB1 containing the miR-129-binding site were cloned into the pmirGLO reporter (Promega, Madison, WI, USA). The mutant type (MUT) of 3’-UTR H19 and 3’-UTR HMGB1 was established by TaKaRa MutanBEST kit (TaKaRa, Japan). Then, the reporter vector and miR-129 were co-transfected into cells via Lipofectamine 2000 (Thermo Fisher Scientific) and cell luciferase activity was tested by the luciferase reporter gene system (Promega) 48 h later.
RNA pull-down assay
The cells were transfected with 200 nM biotinylated miRNA for 24 h and lysed with ice-cold RNA pull-down lysis buffer. The lysate was centrifuged to collect the supernatant which was then packaged (50 mL). The remaining lysate was reacted with streptavidin magnetic beads (Thermo Scientific Fisher, San Jose, CA, USA), RNase-bovine serum albumin and yeast tRNA (both from Sigma-Aldrich). The beads were purified by low-salt and high-salt buffer. Finally, purified by Trizol (Sigma-Aldrich), the bound RNA was utilized to measure H19 level by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) [26].
RT-qPCR
Total RNA was extracted from the Trizol kit (Invitrogen), and the concentration and OD value of the extracted total RNA were measured via a spectrophotometer. For mRNA and lncRNA expression analysis,RNA was reverse transcribed into cDNA with RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific™, USA). PCR was performed on an ABI7500 real-time PCR instrument via using SYBR premix Ex TaqTM II PCR Kit (Takara, Dalian, Liaoning, China). Primers were synthesized by Invitrogen (Table 1). Glyceraldehyde phosphate dehydrogenase (GAPDH) and U6 were the loading controls. 2−ΔΔCt method was utilized to gene expression analysis.
Table 1.
Primer sequence
| Gene | Sequence (5ʹ→3ʹ) |
|---|---|
| MiR-129 | F: CTTTTTGCGGTCTGGGCTTGC |
| R: CTTTTTGCGGTCTGGGCTTGC | |
| U6 | F: GCTTCGGCAGCACATATACTAA |
| R: AACCGCTTCACGAATTTGCGT | |
| H19 | F: TGATGACGGGTGGAGGGGCTA |
| R: TGATGTCGCCCTGTCTGCACG | |
| HMGB1 | F: GCCTCCTTCGGCCTTCTT |
| R: ACAGGCCAGGATGTTCTCCTTT | |
| Bax | F: GGATGCGTCCACCAAGAA |
| R: TCCCGCAGGAAGCCAAT | |
| Bcl-2 | F: CCTTTGTCTAACTCTACCCCC |
| R: CTTTGCCAGTAAATAGCTGATTCCAG | |
| Ki-67 | F: GCAAGCTCACACAAACCTCA |
| R: ACATCTTCTGCGTCCAGCTT | |
| P53 | F: AGAGGAATCCCAAAGTTCCA |
| R: AGAGGAATCCCAAAGTTCCA | |
| GAPDH | F: GGAGCGAGATCCCTCCAAAAT |
| R: GGTTGTTGTCATACTTCTCATGG |
Note: MiR-129, mocroRNA-129; H19, long non-coding RNA H19; HMGB1, high mobility group box-1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; F, forward; R, reverse.
Western blot analysis
Cells were lysed with raidoimmunoprecipitation assay reagent (Beyotime, ShangHai, China) in the presence of protease inhibitors (Sigma, USA) and total protein quantified with the bicinchoninic acid protein assay kit (Beyotime). An appropriate amount of protein was joined to the loading buffer. The protein was denatured and centrifuged, and the supernatant and the loading buffer were taken. The sample with 20 μg was joined to each well, conducted with electrophoresis and electro-blotted onto polyvinylidene difluoride membrane. The membrane was blocked with 50 g/L skim milk powder, incubated with primary antibody HMGB1 (1:10,000; Abcam) and the secondary antibody corresponding to the horseradish peroxidase label, developed with chemiluminescence solution, and analyzed for gray value using Image software.
AD modeling with mice
BALB/c male mice (n = 20), 6 wk old, 22-24 g, were provided by the animal center of Inner Mongolia University (Inner Mongolia, China) and raised in specific pathogen-free animal rooms with constant temperature and humidity, 12-h lighting (08:00–20:00), and enough food and water. Mice were assigned into the sham group and model group (n = 10/group).
An AD mouse model was established [27]. Mice were injected intraperitoneally with pentobarbital sodium (40 mg/kg) [28], after which routine skin preparation and sterilization were performed. Mice were fixed in a brain locator with the anterior and posterior fontanel at the same level. With the anterior fontanel exposed, a small hole was performed with the needle of a 5-mL syringe (Bregma point as the origin, 3.0 mm behind the anterior fontanel, 2.2 mm next to the midline, and 2.8 mm beneath the dura mater). From the hole, 10 μL of aggregated Aβ25-35 (2 mg/ml) was inserted with a 10-μL microsyringe. Aβ25-35 was injected into the right ventricle for 10 min with the needle kept for 10 min. The mice were injected with 4 × 104 U penicillin every day for 3 d to prevent infection. Mice in the sham group were injected with 10 μL 0.9% sodium chloride solution into the ventricles and treated same as the model group. Mice were euthanized to harvest hippocampus.
Statistical analysis
The data were statistically analyzed through SPSS 21.0 (IBM Corp. Armonk, NY, USA) statistical software. All data were expressed as mean ± standard deviation. Student’s t test was used to evaluate significant differences between any two groups of data. One-way analysis of variance (ANOVA) was used for comparison among multiple groups, and the Tukey’s post hoc test was functioned in pairwise comparison after ANOVA analysis. P < 0.05 was considered statistically significant.
Results
Morphology of PC12 cells induced by Aβ25-35
PC12 cells were induced by Aβ25-35, and the changes of PC12 cells treated with different concentrations of Aβ25-35 for 24 h were observed. Microscopic observation revealed that (Figure 1a): normal PC12 cells were spindle-shaped, forming clusters, and cell synapses did not shrink or shortened. There was regular nuclear growth and no nuclear dissolution, or nuclear fragmentation. The cell growth density was evenly distributed. By contrast with the normally grown PC12 cells, as the concentration of Aβ25-35 modeling agent increased, the fusiform shape became irregular and square, the synapses became shorter and smaller, and the cell size shrunk and was unevenly distributed. The nucleus were dissolved and fragmented, and the number of cells was decreased.
Figure 1.
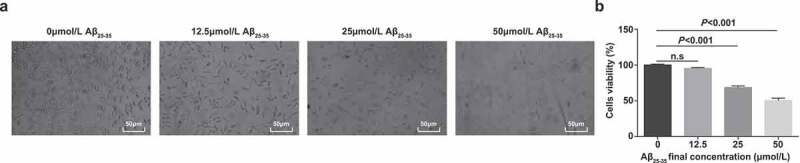
Observation of the morphology of PC12 cells induced by Aβ25-35. A. Observation of the morphology of PC12 cells induced by Aβ25-35 under a microscope; B. The viability of PC12 cells induced by Aβ25-35 tested by MTT assay, N = 3. Data were expressed as mean ± standard deviation. One way ANOVA was functioned for comparison among multiple groups, followed by Tukey's post hoc test
MTT assay detecting cell viability found that (Figure 1b) Aβ25-35 had toxicity and killing effect on PC12 cells. With the increase of the concentration of Aβ25-35 modeling agent, the number of survival cells was declined gradually, and the toxicity and killing effect of Aβ25-35 on the cells were enhanced in a dose-dependent manner. Thus, 25 μmol/L Aβ25-35 was selected to induce PC12 cells.
Elevated H19, HMGB1 and declined miR-129 exhibit in PC12 cells after induction of Aβ25-35
First of all, RT-qPCR was conducted to measure expression of miR-675, miR-106a, miR-194-5p, miR-342-3p, miR-216a-5p, miR-129, miR-148a-3p, miR-29b-3p and miR-93-5p expression and figured out that miR-129 had the largest difference (Supplementary Figure 1). Therefore, miR-129 was selected in the follow-up experiments. H19, miR-129 and HMGB1 levels in PC12 cells induced by 0 μmol/L and 25 μmol/L Aβ25-35 were tested (Figure 2a-c). By contrast with 0 μmol/L Aβ25-35, H19 and HMGB1 expression were elevated, miR-129 expression was decreased in PC12 cells induced by 25 μmol/L Aβ25-35 (all P < 0.05).
Figure 2.
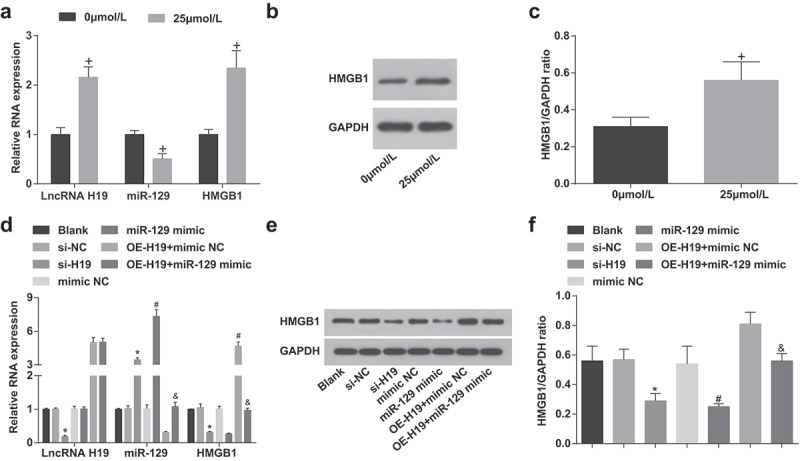
Elevation of H19, HMGB1 and reduction of miR-129 exhibit in PC12 cells after induction of Aβ25-35 A. H19, miR-129, HMGB1 expression in PC12 cells treated with different concentrations of Aβ25-35 detected via RT-qPCR; B. HMGB1 protein bands in PC12 cells treated with different concentrations of Aβ25-35; C. HMGB1 protein expression in PC12 cells treated with different concentrations of Aβ25-35 detected via western blot analysis; D. H19, miR-129, HMGB1 expression in Aβ25-35-treated PC12 cells after transfection detected via RT-qPCR; E. HMGB1 protein bands in Aβ25-35-treated PC12 cells after transfection detected via western blot analysis; F. HMGB1 protein expression in Aβ25-35-treated PC12 cells after transfection detected via western blot analysis. The data in the figure were all measurement data, in the form of mean ± standard deviation. Student’s t test was used to evaluate significant differences between two groups of data. One-way ANOVA was functioned for comparison among multiple groups. The pairwise comparison after ANOVA analysis used Tukey’s post hoc test, N = 3, In the A and C, + vs 0 μmol/L, P < 0.05; In the D and F, * vs the si-NC group, P < 0.05; # vs the mimic NC group; & vs the OE-H19 + mimic NC group, P < 0.05
Silenced H19 increases miR-129 and decreases HMGB1 expression; up-regulated miR-129 reduces HMGB1 expression in Aβ25-35-induced PC12 cells
H19, miR-129 and HMGB1 expression levels in PC12 cells induced by Aβ25-35 were tested (Figure 2d-f). Versus the si-NC group, H19 expression and HMGB1 were reduced while miR-129 expression was increased obviously in Aβ25-35-induced PC12 cells of the si-H19 group (all P < 0.05). With respect to the mimic NC group, miR-129 was elevated while HMGB1 was reduced in Aβ25-35-induced PC12 cells of the miR-129 mimic group (both P < 0.05). By comparison with the OE-H19 + mimic NC group, there were up-regulated miR-129 and downregulated HMGB1 in Aβ25-35-induced PC12 cells of the OE-H19 + miR-129 mimic group (both P < 0.05).
Silenced H19 or elevated miR-129 promote viability and colony formation ability of PC12 cells induced by Aβ25-35
Plenty of assays (MTT and colony formation assay) were functioned to detect the viability and colony formation ability of PC12 cells induced by Aβ25-35 (Figure 3a-c). Elevated viability, proliferation rate and colony formation ability were exhibited in Aβ25-35-induced PC12 cells of the si-H19, the miR-129 mimic and the OE-H19 + miR-129 mimic groups in contrast with the si-NC, the mimic NC, and the OE-H19 + mimic NC groups severally (all P < 0.05).
Figure 3.
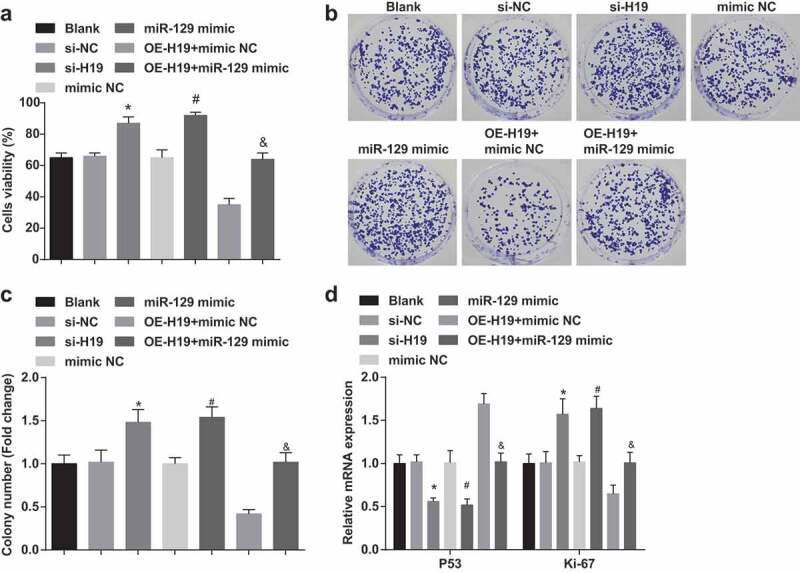
Silence of H19 or elevation of miR-129 facilitate viability and colony formation ability of PC12 cells induced by Aβ25-35. A. The viability of cells detected by MTT assay; B. The cell colony formation ability tested by colony formation assay; C. Quantification results of panel B; D. Ki-67 and P53 mRNA expression in PC12 cells tested by RT-qPCR; The data in the figure were all measurement data, in the form of mean ± standard deviation. One-way ANOVA was functioned for comparison among multiple groups. Tukey’s post hoc test was employed in pairwise comparison after ANOVA analysis. N = 3, * vs the si-NC group, P < 0.05; # vs the mimic NC group, P < 0.05; & vs the OE-H19 + mimic NC group, P < 0.05
P53 and Ki-67 expression in PC12 cells induced by Aβ25-35 were tested by RT-qPCR. We found that (Figure 3d) decreased P53 and elevated Ki-67 expression were found in Aβ25-35-induced PC12 cells of the si-H19, the miR-129 mimic and the OE-H19 + miR-129 mimic groups in contrast with the si-NC, the mimic NC, and the OE-H19 + mimic NC groups severally (all P < 0.05).
Suppressed H19 or elevated miR-129 accelerate cell cycle progression and inhibit apoptosis of PC12 cells induced by Aβ25-35
Flow cytometry has the advantages of accurate analysis, good repeatability and fast analysis efficacy. Therefore, flow cytometry was carried out in this study to examine the cell cycle distribution and apoptosis of PC12 cells induced by Aβ25-35 (Figure 4a-d). Reduced proportion of cells in G0/G1 phase, elevated proportion of cells in S phase, suppressed proportion of apoptosis (all P < 0.05) and not changed proportion of cells in G2/M phase (P > 0.05) all existed in Aβ25-35-induced PC12 cells of the si-H19, the miR-129 mimic and the OE-H19 + miR-129 mimic groups versus the si-NC, the mimic NC, and the OE-H19 + mimic NC groups severally.
Figure 4.
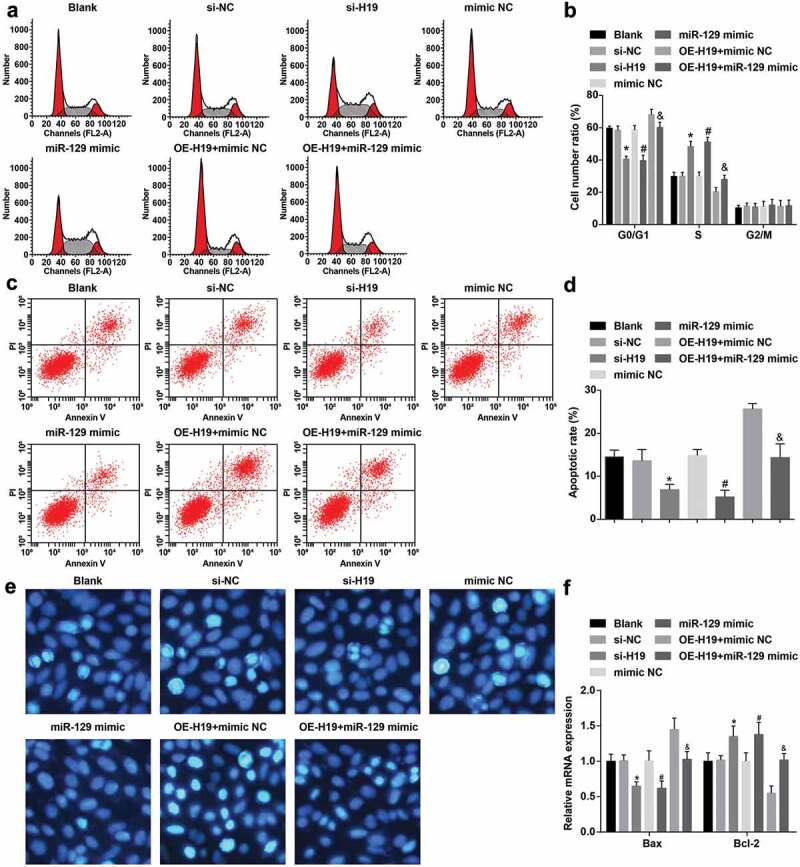
Decreased H19 or elevated miR-129 accelerate cell cycle entry and restrain apoptosis of PC12 cells induced by Aβ25-35. A. Aβ25-35-treated PC12 cell cycle distribution tested by flow cytometry; B. Quantification results of panel A; C. The apoptosis of Aβ25-35-treated PC12 cells tested by flow cytometry; D. Quantification results of panel C; E. The apoptosis of Aβ25-35-treated PC12 cells detected via Hoechst 33,258 staining; F. Bax and Bcl-2 mRNA expression in Aβ25-35-treated PC12 cells tested by RT-qPCR; The data in the figure were all measurement data, in the form of mean ± standard deviation. One-way ANOVA was functioned for comparison among multiple groups. Tukey’s post hoc test was employed in pairwise comparison after ANOVA analysis. N = 3, * vs the si-NC group, P < 0.05; # vs the mimic NC group, P < 0.05; & vs the OE-H19 + mimic NC group, P < 0.05
Hoechst 33258 is a blue fluorescent dye that can penetrate the cell membrane and has low toxicity to cells. Under the fluorescence microscope, the living nucleus show diffuse and uniform fluorescence. When apoptosis occurs, dense granular and massive fluorescence could be seen in the nucleus or cytoplasm. The apoptosis of PC12 cells induced by Aβ25-35 was examined by Hoechst 33258 staining (Figure 4e). PC12 cells in the blank group were seen obvious condensation and rupture, and the apoptotic part could be seen bright blue dots or condensed chromatin. The apoptotic morphology of PC12 cells in the blank, the mimic NC, the OE-H19 + mimic NC groups was not apparently different. By contrast with the si-NC group, a small amount of condensed and ruptured cells were observed, and a bright blue half point or condensed chromatin was observed in the apoptotic portion in PC12 cells in the si-H19 group. Versus the mimic NC group, the nuclear apoptotic morphology of PC12 cells in the miR-129 mimic group was apparently reduced, with a few bright blue dots or condensed chromatin. With the OE-H19 + mimic NC group in contrast, PC12 cells could be condensed and ruptured in a small amount, and a little apoptotic part could be seen with a bright blue half point or condensed chromatin in the OE-H19 + miR-129 mimic group.
Furthermore, detecting Bax and Bcl-2 expression can further explain the impact on apoptosis at the molecular level. Bax and Bcl-2 expression in Aβ25-35-treated PC12 cells were tested by RT-qPCR (figure 4f). A reduction of Bax mRNA expression and an elevation of Bcl-2 mRNA expression were manifested in Aβ25-35-induced PC12 cells of the si-H19, the miR-129 mimic and the OE-H19 + miR-129 mimic groups in contrast with the si-NC, the mimic NC, and the OE-H19 + mimic NC groups severally (all P < 0.05).
Silenced H19 or elevated miR-129 reduce ROS and up-regulate MMP in PC12 cells induced by Aβ25-35
When detecting ROS and measuring MMP in PC12 cells induced by Aβ25-35, the results indicated that (Figure 5 a,b) decreased ROS and elevated MMP exhibited in Aβ25-35-induced PC12 cells of the si-H19, the miR-129 mimic and the OE-H19 + miR-129 mimic groups in contrast with the si-NC, the mimic NC, and the OE-H19 + mimic NC groups severally (all P < 0.05).
Figure 5.
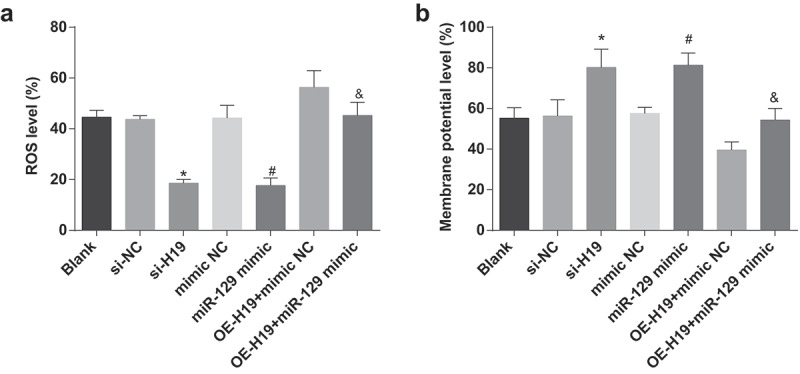
Reduction of H19 or elevation of miR-129 decrease ROS and elevate MMP expression in PC12 cells induced by Aβ25-35. A. ROS in Aβ25-35-treated PC12 cells after transfection; B. MMP in Aβ25-35-treated PC12 cells after transfection. The data in the figure were all measurement data, in the form of mean ± standard deviation. One-way ANOVA was functioned for comparison among multiple groups. Tukey’s post hoc test was employed in pairwise comparison after ANOVA analysis. N = 3, * vs the si-NC group, P < 0.05; # vs the mimic NC group, P < 0.05; & vs the OE-H19 + mimic NC group, P < 0.05
Down-regulated H19 or up-regulated miR-129 inhibit oxidative stress in Aβ25-35 induced PC12 cells
Through testing MDA, SOD, and CAT in PC12 cells induced by Aβ25-35, the results suggested that (Figure 6a-c) SOD and CAT expression were elevated while MDA expression was decreased in Aβ25-35-induced PC12 cells of the si-H19, the miR-129 mimic and the OE-H19 + miR-129 mimic groups in contrast with the si-NC, the mimic NC, and the OE-H19 + mimic NC groups severally (all P < 0.05).
Figure 6.
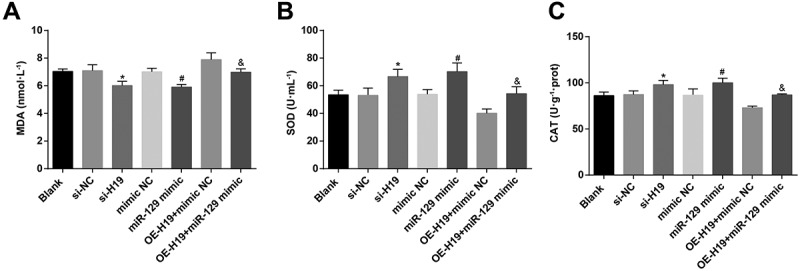
Inhibited H19 or up-regulated miR-129 elevate SOD and CAT and reduce MDA in Aβ25-35 induced PC12 cells. A. MDA in Aβ25-35-treated PC12 cells after transfection; B. SOD activity in Aβ25-35-treated PC12 cells after transfection; C. CAT expression in Aβ25-35-treated PC12 cells after transfection. The data in the figure were all measurement data, in the form of mean ± standard deviation. One-way ANOVA was functioned for comparison among multiple groups. Tukey’s post hoc test was employed in pairwise comparison after ANOVA analysis. N = 3, * vs the si-NC group, P < 0.05; # vs the mimic NC group, P < 0.05; & vs the OE-H19 + mimic NC group, P < 0.05
H19 binds with miR-129
To explore the mechanism of action of H19, we first analyzed it by online analysis (http://lncatlas.crg.eu/) (Figure 7a) and found that H19 was mainly distributed in the cytoplasm, which was further verified in RNA-FISH assay (Figure 7b). Through the RNA22 website (https://cm.jefferson.edu/rna22/Precomputed/), it was indicated that H19 could bind to miR-129 (Figure 7c). As revealed in the dual-luciferase reporter gene assay, in contrast with the mimic NC group, the luciferase activity of the WT-H19 was decreased in the miR-129 mimic group (P < 0.05), indicating a binding relationship between H19 and miR-129 (Figure 7d). RNA pull-down assay was implemented to verify that H19 could bind with miR-129. The results suggested that the enrichment level of H19 was elevated in the Bio-miR-129-WT group (P < 0.05) by contrast with that in the Bio-NC group (P < 0.05) (Figure 7e). These results emerged that H19 could bind with miR-129, thereby affecting miR-129 expression.
Figure 7.
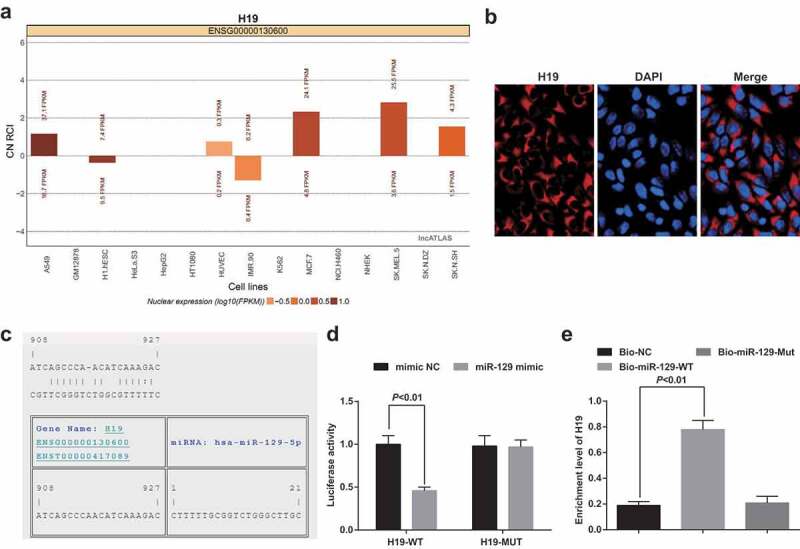
H19 competitively regulates miR-129. A. H19 subcellular localization predicted via online analysis site; B. H19 subcellular localization verified by FISH assay; C. The binding site of H19 to miR-129 predicted via RNA22 site; D. The binding of H19 to miR-129 verified via luciferase activity assay; E. The enrichment of miR-129 on H19 tested via RNA pull-down assay; The data in the figure were all measurement data, in the form of mean ± standard deviation. Student’s t test was used to evaluate significant differences between two groups of data. One-way ANOVA was functioned for comparison among multiple groups, followed by Tukey’s post hoc test. N = 3
HMGB1 is a target gene of miR-129
The target site of HMGB1 binding to the corresponding miR-129 was determined by the online prediction software Target Scan. The sequence of the 3ʹ-UTR region of HMGB1 mRNA binding to miR-129 is shown in Figure 8a. The detection results manifested that (Figure 8b) miR-129 mimic had no effect on the luciferase activity of the HMGB1-MUT plasmid (P > 0.05), but the luciferase activity in the HMGB1-WT plasmid was apparently reduced (P < 0.05).
Figure 8.

The targeting relationship between HMGB1 and miR-129. A. The target site for binding of HMGB1 and the corresponding miR-129 predicted via Target Scan; B. Detection results of dual-luciferase reporter gene assay. The data represented the mean ± standard deviation of three independent experiments. Student’s t test was used to evaluate significant differences between two groups of data. N = 3.* vs the mimic NC group, P < 0.05
H19 and HMGB1 are elevated and miR-129 is decreased in AD mice
Finally, in vivo experiments were conducted to test H19, miR-129 and HMGB1 expression in brain tissues of AD mice. It was suggested that compared with the sham group, H19 and HMGB1 expression were elevated while miR-129 expression was reduced in brain tissues of AD mice (Figure9 a,b).
Figure 9.
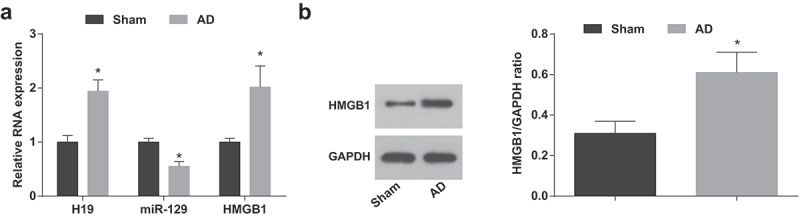
H19 and HMGB1 are elevated and miR-129 is suppressed in AD mice. A. H19, miR-129 and HMGB1 expression in brain tissues of AD mice via RT-qPCR; B. HMGB1 protein expression in brain tissues of AD mice via Western blot analysis. The two-group comparison was performed by independent sample t test for statistical analysis. n = 10. * vs the sham group, P < 0.05
Discussion
AD frequently manifests as dementia [29]. Much studies focus on the important role of lncRNAs in AD. The goal of the study was for the research of the mechanism of H19 in proliferation and apoptosis of PC12 cells induced by Aβ25-35 in AD with modulation of miR-129 and HMGB1.
At first, elevated H19 showed up in PC12 cells after induction of Aβ25-35 in AD. Similar to our study, H19 is clearly up-regulated in the latent period of TLE [9]. Meanwhile, H19 also demonstrated to be overexpressed in Schwann cells isolated from peripheral nerves in the peripheral neuropathic pain [8]. Analyzed by various assays, silenced H19 was proved to promote viability, colony formation ability, and cycle entry, inhibit apoptosis, and decrease oxidative stress in Aβ25-35 induced PC12 cells. Similar results were gained in the following studies. For instance, a study has indicated that depressed H19 restrains lung carcinoma cell proliferation and overexpressed H19 has a contrast effect [30]. Another study has emerged that H19 overexpression strengthens lung cancer cell proliferation by a similar mechanism in colorectal cancer [31]. Evidences have shown that H19 plays functional roles in neuronal diseases, such as Parkinson’s disease [32] while the mechanism of H19 in AD is not revealed. Additionally, the observation in the study was that H19 acted as a ceRNA to absorb miR-129, which shall be further explored in future studies.
The current study found that declined miR-129 exhibited in PC12 cells after induction of Aβ25-35 in AD. It has been proved that highly expressed miR-129-5p in rats will inhibit neuronal apoptosis and reduce the inflammatory reaction [13]. Another finding was that elevated miR-129 facilitated viability, colony formation ability, and cycle entry, inhibited apoptosis, and decreased oxidative stress in Aβ25-35 induced PC12 cells. A prior study has provided evidence highlighting that miR-129-5p up-regulation could restrain neuronal apoptosis, promote proliferation, suppress apoptosis in neuronal cells in AD [13]. What’s more, this is consistent with our finding that elevated miR-129 mediated by dexmedetomidine resulted in declined neuronal apoptosis in Aβ-injected mice [15]. In this present study, the result also indicated that HMGB1 was a target gene of miR-129. In accordance with the present results, previous studies have demonstrated that miR-129-5p which could specifically bind to HMGB1-3ʹUTR, and depress HMGB1 activity [33]. Moreover, down-regulation of miR-129-5p could directly facilitate the expression of HMGB1 [34]. Furthermore, it is also demonstrated that enhanced miR-129-5p level depresses HMGB1 [35]. A major finding of this study was that elevated HMGB1 occurred in PC12 cells after induction of Aβ25-35 in AD. These data show that elevated HMGB1 stimulates memory abnormalities, manifesting that contributing to memory deficits under various neurological and psychiatric conditions is linked with the increased HMGB1 levels, such as AD [36]. Meanwhile, HMGB1 activation is verified in AD [19]. What’s more, another study has also exhibited that suppression of HMGB1 has been manifested to be effective for AD treatment [37].
In conclusion, our study performs that silenced H19 and up-regulated miR-129 facilitate viability and restrain apoptosis of PC12 cells induced by Aβ25-35 in a cellular model of AD. These results offer promising insight into the function of H19 in the progression of AD, and might allow for the pursuit of novel therapeutics and diagnostic markers for AD. More researches should be undertaken to elucidate the mechanism of HMGB1 in AD more clearly.
Supplementary Material
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper.
Funding Statement
This work was supported by the Inner Mongolia Natural Science Foundation [No. 2018MS08127], National Natural Science Foundation of China [No. 81660720] and Program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region [No. NJYT −20-B24].
Disclosure statement
The authors declare that they have no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Saeedi M, Rastegari A, Hariri R, et al. Design and synthesis of novel arylisoxazole-chromenone carboxamides: investigation of biological activities associated with Alzheimer’s disease. Chem Biodivers. 2020;17. DOI: 10.1002/cbdv.201900746. [DOI] [PubMed] [Google Scholar]
- [2].Wu J, Zhang X, Azhati G, et al. Retinal microvascular attenuation in mental cognitive impairment and Alzheimer’s disease by optical coherence tomography angiography. Acta Ophthalmol. 2020;98. DOI: 10.1111/aos.14381. [DOI] [PubMed] [Google Scholar]
- [3].Tseng HJ, Lin M-H, Shiao Y-J, et al. Synthesis and biological evaluation of acridine-based histone deacetylase inhibitors as multitarget agents against Alzheimer’s disease. Eur J Med Chem. 2020;192:112193. [DOI] [PubMed] [Google Scholar]
- [4].Shi Y, Liu H, Yang C, et al. Transcriptomic analyses for identification and prioritization of genes associated with Alzheimer’s disease in humans. Front Bioeng Biotechnol. 2020;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sen S , et al. Neurodegeneration in Alzheimer’s disease and glaucoma: overlaps and missing links. Eye (Lond). 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu Y, Gong Y, Xie W, et al. Microbubbles in combination with focused ultrasound for the delivery of quercetin-modified sulfur nanoparticles through the blood brain barrier into the brain parenchyma and relief of endoplasmic reticulum stress to treat Alzheimer’s disease. Nanoscale. 2020;12(11):6498–6511. [DOI] [PubMed] [Google Scholar]
- [7].Li X, Luo S, Zhang J, et al. lncRNA H19 alleviated myocardial I/RI via suppressing miR-877-3p/Bcl-2-mediated mitochondrial apoptosis. Mol Ther Nucleic Acids. 2019;17:297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Iwasaki H, Sakai A, Maruyama M, et al. Increased H19 long non-coding RNA expression in Schwann cells in peripheral neuropathic pain. J Nippon Med Sch. 2019;86(4):215–221. [DOI] [PubMed] [Google Scholar]
- [9].Han CL, Ge M, Liu Y-P, et al. Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell Death Dis. 2018;9(6):617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vijayan M, Reddy PH.. Non-coding RNAs based molecular links in type 2 diabetes, ischemic stroke, and vascular dementia. J Alzheimers Dis. 2020;75(2):353–383. [DOI] [PubMed] [Google Scholar]
- [11].Gao Y, Feng B, Han S, et al. MicroRNA-129 in human cancers: from tumorigenesis to clinical treatment. Cell Physiol Biochem. 2016;39(6):2186–2202. [DOI] [PubMed] [Google Scholar]
- [12].Wu DM, Zhang Y-T, Lu J, et al. Effects of microRNA-129 and its target gene c-Fos on proliferation and apoptosis of hippocampal neurons in rats with epilepsy via the MAPK signaling pathway. J Cell Physiol. 2018;233(9):6632–6643. [DOI] [PubMed] [Google Scholar]
- [13].Zeng Z, Liu Y, Zheng W, et al. MicroRNA-129-5p alleviates nerve injury and inflammatory response of Alzheimer’s disease via downregulating SOX6. Cell Cycle. 2019;18(22):3095–3110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [14].Li Z, Chen Q, Liu J, et al. Physical exercise ameliorates the cognitive function and attenuates the neuroinflammation of Alzheimer’s disease via miR-129-5p. Dement Geriatr Cogn Disord. 2020;49(2):163–169. [DOI] [PubMed] [Google Scholar]
- [15].Sun W, Zhao J, Li C. Dexmedetomidine provides protection against hippocampal neuron apoptosis and cognitive impairment in mice with Alzheimer’s disease by mediating the miR-129/YAP1/JAG1 axis. Mol Neurobiol. 2020;57(12):5044–5055. [DOI] [PubMed] [Google Scholar]
- [16].Yang Y, Huang J-Q, Zhang X, et al. MiR-129-2 functions as a tumor suppressor in glioma cells by targeting HMGB1 and is down-regulated by DNA methylation. Mol Cell Biochem. 2015;404(1–2):229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Davis HM, Valdez S, Gomez L, et al. High mobility group box 1 protein regulates osteoclastogenesis through direct actions on osteocytes and osteoclasts in vitro. J Cell Biochem. 2019;120(10):16741–16749. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fujita K, Motoki K, Tagawa K, et al. HMGB1, a pathogenic molecule that induces neurite degeneration via TLR4-MARCKS, is a potential therapeutic target for Alzheimer’s disease. Sci Rep. 2016;6(1):31895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nan K, Han Y, Fang Q, et al. HMGB1 gene silencing inhibits neuroinflammation via down-regulation of NF-kappaB signaling in primary hippocampal neurons induced by Abeta25-35. Int Immunopharmacol. 2019;67:294–301. [DOI] [PubMed] [Google Scholar]
- [20].Shao X, Ma W, Xie X, et al. Neuroprotective effect of tetrahedral DNA nanostructures in a cell model of Alzheimer’s disease. ACS Appl Mater Interfaces. 2018;10(28):23682–23692. [DOI] [PubMed] [Google Scholar]
- [21].Wei D, et al. Synthesis, characterization, antioxidant activity and neuroprotective effects of selenium polysaccharide from Radix hedysari. Carbohydr Polym. 2015;125:161–168. [DOI] [PubMed] [Google Scholar]
- [22].Xi YD, Zhang -D-D, Ding J, et al. Genistein Inhibits Abeta25-35-induced synaptic toxicity and regulates CaMKII/CREB pathway in SH-SY5Y cells. Cell Mol Neurobiol. 2016;36(7):1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yan H, et al. Upregulation of miR-183-5p is responsible for the promotion of apoptosis and inhibition of the epithelial-mesenchymal transition, proliferation, invasion and migration of human endometrial cancer cells by downregulating Ezrin. Int J Mol Med. 2018;42(5):2469–2480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [24].Hu Y, Deng C, Zhang H, et al. Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/beta-catenin signaling pathway in bladder cancer. Oncotarget. 2017;8(55):94554–94568. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [25].David D, Alfredo P, Eduardo R, et al. Flow cytometry enumeration of apoptotic cancer cells by apoptotic rate. Methods Mol Biol. 2015;1219:11–20. [DOI] [PubMed] [Google Scholar]
- [26].Li D, Liu Y, Gao W, et al. LncRNA HCG11 inhibits adipocyte differentiation in human adipose-derived mesenchymal stem cells by sponging miR-204-5p to upregulate SIRT1. Cell Transplant. 2020;29:963689720968090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhu J, Chen X, Song Y, et al. Deficit of RACK1 contributes to the spatial memory impairment via upregulating BECLIN1 to induce autophagy. Life Sci. 2016;151:115–121. [DOI] [PubMed] [Google Scholar]
- [28].Lv X, Wang Z-M, Huang S-D, et al. Emulsified isoflurane preconditioning reduces lung injury induced by hepatic ischemia/reperfusion in rats. Int J Med Sci. 2011;8(5):353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen Y, Zhang J, Zhang T, et al. Meditation treatment of Alzheimer disease and mild cognitive impairment: A protocol for systematic review. Medicine (Baltimore). 2020;99(10):e19313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liao S, Yu C, Liu H, et al. Long non-coding RNA H19 promotes the proliferation and invasion of lung cancer cells and regulates the expression of E-cadherin, N-cadherin, and vimentin. Onco Targets Ther. 2019;12:4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhao Y, Feng C, Li Y, et al. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem. 2019;460(1–2):1–8. [DOI] [PubMed] [Google Scholar]
- [32].Jiang J, Piao X, Hu S, et al. LncRNA H19 diminishes dopaminergic neuron loss by mediating microRNA-301b-3p in Parkinson’s disease via the HPRT1-mediated Wnt/beta-catenin signaling pathway. Aging (Albany NY). 2020;12(10):8820–8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu AH, Wu YT, Wang YP. MicroRNA-129-5p inhibits the development of autoimmune encephalomyelitis-related epilepsy by targeting HMGB1 through the TLR4/NF-kB signaling pathway. Brain Res Bull. 2017;132:139–149. [DOI] [PubMed] [Google Scholar]
- [34].Wang S, Chen Y, Yu X, et al. miR-129-5p attenuates cell proliferation and epithelial mesenchymal transition via HMGB1 in gastric cancer. Pathol Res Pract. 2019;215(4):676–682. [DOI] [PubMed] [Google Scholar]
- [35].Li XQ, Chen F-S, Tan W-F, et al. Elevated microRNA-129-5p level ameliorates neuroinflammation and blood-spinal cord barrier damage after ischemia-reperfusion by inhibiting HMGB1 and the TLR3-cytokine pathway. J Neuroinflammation. 2017;14(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mazarati A, Maroso M, Iori V, et al. High-mobility group box-1 impairs memory in mice through both toll-like receptor 4 and Receptor for Advanced Glycation End Products. Exp Neurol. 2011;232(2):143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nishibori M, Mori S, Takahashi HK. Anti-HMGB1 monoclonal antibody therapy for a wide range of CNS and PNS diseases. J Pharmacol Sci. 2019;140(1):94–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


