Aplastic anemia (AA) is bone marrow failure characterized by a hypocellular marrow and peripheral pancytopenia. Immunosuppressive therapy (IST) results in hematologic recovery in the majority, decreased short term cytopenia related complications, and increased survival. However, clonal evolution to a secondary myeloid malignancy is a major complication in long-term survivors.1 Clonal evolution occurs in 10-15% of severe aplastic anemia (SAA) patients after IST.2 Most high risk clonal evolution is associated with complete or partial loss of chromosome 7, which is also frequent in other marrow failure syndromes.3 Previous studies have found older age, multiple rounds of IST, and severity of cytopenia as risk factors for clonal evolution.2,3
Shorter telomere length at diagnosis of AA and accelerated telomere attrition preceding the malignant transformation are associated with chromosome 7 clonal evolution, suggesting genomic instability as a possible mechanism. 1 Frank myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) without chromosome 7 involvement is much less common.3-5 Mutations in myeloid neoplasia related genes are present in 20-30% of patients with SAA.6,7 Excluding PIGA mutations, the most frequently affected genes are epigenetic regulators: ASXL1, DNMT3A and BCOR. The presence of unfavorable somatic mutations as a group is associated with a higher risk of clonal evolution and poorer survival.7,8 Surprisingly, somatic mutations were not frequently detected in clonal evolution to monosomy 7,9 including in patients treated with eltrombopag (EPAG).10
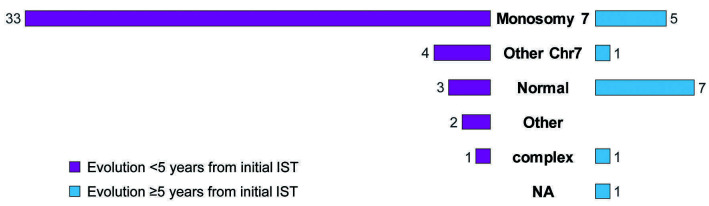
We retrospectively assessed all subjects with SAA treated at the NIH Clinical Center on Hematology Branch protocols, with either horse- anti thymocyte globulin (ATG), rabbit-ATG or alemtuzumab based regimens for treatment-naïve or relapsed/refractory disease from 1989 to 2019. A total of 666 subjects were identified, of whom 96 had clonal evolution. The definition of clonal evolution was consistent across protocols. Clonal evolution was defined as an acquisition of new cytogenetic abnormalities with or without morphologic evidence of a myeloid malignancy. It was considered a high-risk clonal evolution when there was a diagnosis of a myeloid malignancy, an isolated acquisition of chromosome 7 abnormality or complex karyotype. High-risk evolution was noted in 58 of 96 subjects (Figure 1). Fifteen of these 58 subjects had evolved at 5 years or later after initial IST treatment. Among these 15 patients, eight subjects evolved with cytogenetic abnormalities and seven had MDS/AML with normal karyotype. We further investigated whether the late evolution to MDS/AML in the absence of cytogenetic changes was associated with somatic mutations in myeloid genes. Three subjects evolved at outside institutions and were excluded from the analysis due to insufficient data. One subject with loss of Y, a common age-related cytogenetic abnormality, was included.
In this case series, we report four cases of late occurring MDS/AML without chromosome 7 abnormalities. Patients were screened for mutations in genes recurrently mutated in myeloid malignancies by next generation sequencing (NGS) at the time of clonal evolution. Detected variants were confirmed and tracked back in earlier serial samples by digital droplet PCR (ddPCR).
All patients were evaluated with clinic visits, bone marrow examinations, and cytogenetics, as dictated by treatment protocols No NCT number: Rosenfeld, Blood. 1995: NCT00001964 and NCT01623167. All patients gave written informed consent to inclusion in the respective studies. Telomere length (TL) was measured by flowfluorescence in situ hybridization (FISH) at a commercial laboratory (Repeat Diagnostics) or by Southern blotting at the time of SAA diagnosis. The details about NGS methods are included in the Online Supplementary Appendix.
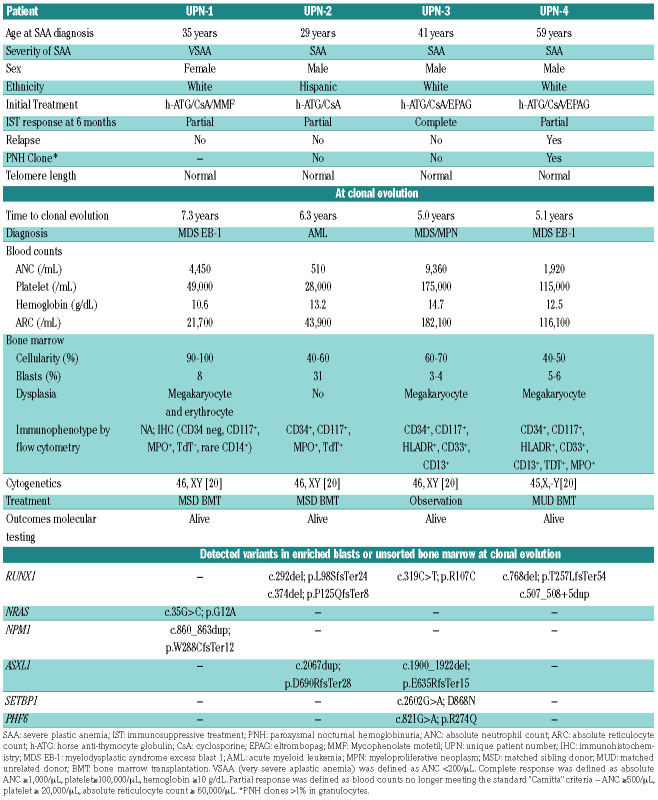
The median age of SAA diagnosis in the four included subjects was 38 years (range: 29-59; Table 1). All four subjects had normal TL at SAA diagnosis. At 6 months, three subjects had achieved partial response and one a complete response (CR) to IST. Only unique patient number (UPN)-4 relapsed 7 months after treatment and required 2 years of additional cyclosporine and EPAG until CR was achieved. This subject also received eculizumab treatment for a large glycosylphosphatidylinositol (GPI) deficient granulocyte clone.
At evolution to myeloid malignancy, median time from initial IST was 5.7 years (range: 5-7.3 years). UPN-1 had pancytopenia and UPN-2 had progressive thrombocytopenia at clonal evolution, but UPN-3 and UPN-4 were diagnosed on protocol-mandated bone marrow evaluations with minimal to no change in their blood counts. Cytogenetics were normal in all except UPN-4, who had loss of chromosome Y present a year prior to evolution.
Figure 1.
Cytogenetic abnormalities in high-risk clonal evolution stratified by time to evolution from initial immunosuppressive treatment for severe aplastic anemia. There were 58 high-risk clonal evolutions in this large cohort of 666 subjects with severe aplastic anemia (SAA) diagnosis treated with immunosuppressive treatment (IST). Majority occurred before 5 years and the most common cytogenetic abnormalities were in chromosome 7 including aneuploidy, partial loss/deletion, or inversion. Chr7: chromosome 7; NA: not available.
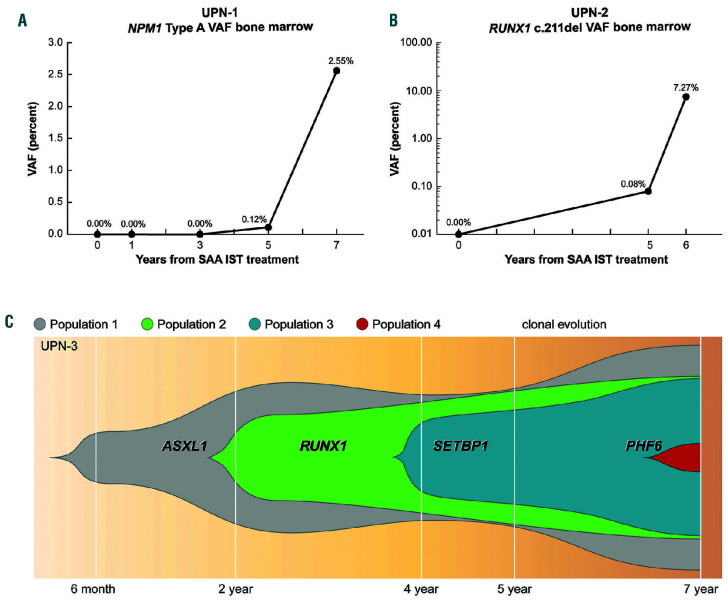
Except for UPN-2, all subjects had less than 10% blasts at evolution. In order to determine the presence of mutations within the malignant clone, we performed NGS on UPN-1, UPN-2, and UPN-4 samples from the time of myeloid malignancy diagnosis that had been enriched by flow cytometry sorting based on blast immunophenotype (Table 1; Online Supplementary Table S1A). For UPN- 1, NGS of CD117+ peripheral blood mononuclear cells (PBMC) at clonal evolution demonstrated mutations in NPM1 (Type A insertion) and NRAS. A custom ddPCR assay for the NPM1 mutation detected the variant in unsorted bone marrow taken 128 and 864 days prior to clonal evolution but not from samples collected prior to that (Figure 2A). In UPN-2, two variants in RUNX1 and one in ASXL1 were detected in sorted CD117+ from bone marrow mononuclear cells at clonal evolution. A custom ddPCR assay for the RUNX1 c.292del mutation detected the mutation in unsorted bone marrow from clonal evolution at variant allele frequency (VAF) of 7% and also 497 days prior to clonal evolution at VAF <1%, but not in a bone marrow sample 2163 days prior to clonal evolution (Figure 2B).
Table 1.
Patient characteristics and details of clonal evolution.
UPN-3 and UPN-4 had clinical NGS testing at clonal evolution. For UPN-3, clinical NGS reported an ASXL1 variant at 6 months post-IST with successive acquisition of RUNX1, SETBP1, and PHF6 variants at 2, 5, and 7 years post-IST respectively (Online Supplementary Table S1B). In order to elucidate the clonal architecture in this subject, we employed a microfluidics-based, single-cell DNA sequencing assay on a sample from the time of malignancy diagnosis which demonstrated that these mutations were all present within the same clone derived from an initial ASXL1 mutated clonal population (Figure 2C). While no somatic variants were reported by clinical NGS analysis in UPN-4 at the time of clonal evolution, research NGS of CD34+ sorted PBMC from this timepoint demonstrated two RUNX1 mutations (Table 1). A custom ddPCR assay for one of these variants (p.Thr257LeufsTer54) confirmed detection at VAF <2% in unsorted bone marrow collected at the time of clonal evolution but not in a marrow sample from 1 year prior.
UPN-1, UPN-2 and UPN-4 underwent allogeneic bone marrow transplantation and all three were alive and disease- free at last follow-up. UPN-3 was followed expectantly with stable counts but a repeat bone marrow 2 years after clonal evolution and 7 years from initial IST revealed morphologic progression with increased dysplasia, presence of trisomy 21, and acquisition of PHF6 somatic mutation.
Clonal evolution after IST for SAA has a varied clinical course. Although high risk evolution with chromosome 7 aneuploidy with or without morphologic evidence of myeloid malignancy is the most common, fewer SAA patients evolve to MDS/AML with normal cytogenetics or without chromosome 7 abnormalities. Isolated chromosome abnormalities, such as deletion 13q and trisomy 8, are considered low risk because patients appear to have better overall survival and less progression to AML.11 Late clonal evolution to MDS/AML without typical cytogenetic abnormalities after IST for SAA occurs in a very small proportion of patients.
Much investigation has been done to understand the underlying etiology of clonal evolution associated with monosomy 7 but a deeper understanding of other forms of high-risk evolution is lacking. We report here the first case series to our knowledge describing clinical and genetic features of SAA patients developing MDS/AML without chromosome 7 abnormalities, arising more than 5 years after IST. In all four patients, we identified molecular alterations in genes recurrently mutated in myeloid malignancies: RUNX1, NRAS, NPM1, ASXL1, SETBP1, PHF6.12 Three subjects had at least one RUNX1 variant, for which the reported prevalence is 10% in de novo AML/MDS or therapy related myeloid neoplasm.12 Somatic RUNX1 mutations have also been described in myeloid malignancies arising from inherited marrow failure including Fanconi anemia (20%) and congenital neutropenia (64%).13,14 Accelerated telomere attrition correlates with monosomy 7 transformation,9 but acquisition of somatic mutations, specifically RUNX1, may be implicated in late high risk clonal evolution without chromosome 7 aberration. Previously, ASXL1 has been associated with higher risk of myeloid malignancy with abnormal cytogenetics.8
Figure 2.
Detection of mutations in three severe aplastic anemia subjects prior to clonal evolution. (A) Presence of NPM1 mutation detected by digital droplet PCR (ddPCR) at, and 2 years prior to, the diagnosis of clonal evolution in unique patient number (UPN)-1; (B) RUNX1 variant was found using ddPCR at, and 1 year before, the diagnosis of clonal evolution in UPN-2; (C) single-cell DNA sequencing at the time of clonal evolution in UPN-3 demonstrates that the successive acquisition of mutations over a 7-year period, including 2 years after the diagnosis of clonal evolution to a myeloid malignancy were all within the same clonal population. SAA: severe aplastic anemia; VAF: variant allele frequency.
Whether clonal evolution to MDS/AML is a result of genomic instability present at diagnosis or developing after IST has not been elucidated. We were able to detect somatic variants present in bone marrow samples from 1-2 years prior to clonal evolution but not at initial SAA diagnosis. Given the rare nature of late evolution to myeloid malignancy without chromosomal abnormalities, and the current limitations of NGS for de novo discovery of MDS/AML variants,15 it is not known if sequential molecular monitoring would be clinically useful for early detection of evolving myeloid malignancy. The utility of early interventions designed to prevent development of myeloid malignancy in high-risk individuals is currently unknown. Approximately 10% of SAA patients treated with IST in this large cohort developed high-risk clonal evolution, supporting the importance of regular long-term clinical follow-up.
Supplementary Material
Funding Statement
Funding: this research was supported in part by the Intramural Research Program of the NIH and NHLBI.
References
- 1.Young NS. Aplastic anemia. N Engl J Med. 2018;379(17):1643-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheinberg P., Young NS. How I treat acquired aplastic anemia. Blood. 2012;120(6):1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Li X, Ge M, et al. Long-term follow-up of clonal evolutions in 802 aplastic anemia patients: a single-center experience. Ann Hematol. 2011;90(5):529-537. [DOI] [PubMed] [Google Scholar]
- 4.Maciejewski JP, Balasubramanian SK. Clinical implications of somatic mutations in aplastic anemia and myelodysplastic syndrome in genomic age. Hematology Am Soc Hematol Educ Program. 2017; 2017(1):66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Planque MM, Bacigalupo A, Wursch A, et al. Long-term followup of severe aplastic anaemia patients treated with antithymocyte globulin. Severe Aplastic Anaemia Working Party of the European Cooperative Group for Bone Marrow Transplantation (EBMT). Br J Haematol. 1989;73(1):121-126. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa S. Clonal hematopoiesis in acquired aplastic anemia. Blood. 2016;128(3):337-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulasekararaj AG, Jiang J, Smith AE., et al. , Somatic mutations identify a subgroup of aplastic anemia patients who progress to myelodys plastic syndrome. Blood. 2014;124(17):2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, Ge M, Lu S, et al. Mutations of ASXL1 and TET2 in aplastic anemia. Haematologica. 2015;100(5):e172-e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumitriu B, Feng X, Townsley DM, et al. Telomere attrition and candidate gene mutations preceding monosomy 7 in aplastic anemia. Blood. 2015;125(4):706-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler T, Fan X, Cooper J, et al. Treatment optimization and genomic outcomes in refractory severe aplastic anemia treated with eltrombopag. Blood. 2019;133(24):2575-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maciejewski JP, Risitano A, Sloand EM, Nunez O, Young NS, et al. Distinct clinical outcomes for cytogenetic abnormalities evolving from aplastic anemia. Blood. 2002;99(9):3129-3135. [DOI] [PubMed] [Google Scholar]
- 12.Haferlach T, Nagata Y, Grossman V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quentin S, Cuccuini W, Ceccaldi R, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117(15):e161-e170. [DOI] [PubMed] [Google Scholar]
- 14.Skokowa J, Steinemann D, Katsman-Kuipers JE, et al. Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood. 2014; 123(14):2229-2237. [DOI] [PubMed] [Google Scholar]
- 15.Ghannam J, Dillon LW, Hourigan CS. Next-generation sequencing for measurable residual disease detection in acute myeloid leukaemia. Br J Haematol. 2020;188(1):77-85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.