ABSTRACT
Background
Human milk contains a diverse community of bacteria believed to play a role in breast health and inoculation of the infant's gastrointestinal tract. The role of maternal nutrition and infant feeding practices on the human milk microbiota remains poorly understood.
Objective
Our aim was to explore the associations between maternal diet (delivery to 3 mo postpartum), infant feeding practices, and the microbial composition and predicted function in milk from women with varied metabolic status.
Methods
This was an exploratory analysis of a previously completed prospective cohort study of women with varying degrees of gestational glucose intolerance (NCT01405547). Milk samples (n = 93 mothers) were collected at 3 mo postpartum. Maternal dietary information (validated food-frequency questionnaire) and infant feeding practices (human milk exclusivity, frequency of direct breastfeeding per day) were collected. V4-16S ribosomal RNA gene sequencing (Illumina MiSeq) was conducted to determine microbiota composition.
Results
Intake of polyunsaturated fat [β estimate (SE): 0.036 (0.018), P = 0.047] and fiber from grains [0.027 (0.013), P = 0.048] were positively associated with ɑ-diversity (Shannon index) of human milk. Overall microbial composition of human milk clustered based on human milk exclusivity (weighted UniFrac R2 = 0.034, P = 0.015; Bray-Curtis R2 = 0.041, P = 0.007), frequency of direct breastfeeding per day (Bray-Curtis R2 = 0.057, P = 0.026), and maternal fiber intake from grains (Bray-Curtis R2 = 0.055, P = 0.040). Total fiber, fiber from grains, dietary fat, and infant feeding practices were also associated with a number of differentially abundant taxa. The overall composition of predicted microbial functions was associated with total fiber consumption (Bray-Curtis R2 = 0.067, P = 0.036) and human milk exclusivity (Bray-Curtis R2 = 0.041, P = 0.013).
Conclusions
Maternal consumption of fiber and fat, as well as mother's infant feeding practices, are important determinants of the human milk microbiota. Understanding whether these microbial changes impact an infant's overall health and development requires future study.
Keywords: human milk, milk microbiota, gastrointestinal microbiota, maternal nutrition, direct breastfeeding
See corresponding commentary on page 278.
Introduction
It is now known that human milk contains an abundant supply of bacteria, much of it living, ranging from 103 to 106 bacterial cells/mL, depending on whether culture-based methods or molecular methods are used to assess (1–3). These bacteria, some of which are naturally occurring probiotics, are believed to play a part in human health, including their involvement in breast health, as well as in colonizing the neonate's gastrointestinal (GI) tract, orchestrating the development of the infant's GI maturation, and ensuring appropriate tolerization of the naïve immune system (4–8). Several studies have shown that the infant's GI microbiota is associated with a number of both short- and long-term health outcomes, including diarrhea, respiratory tract infection, asthma, inflammatory bowel disease, body size, and metabolic syndrome (9–12).
Despite the colonization potential of microbiota in human milk, factors associated with its microbial composition remain underinvestigated. Of the few available studies, maternal body size, mode of delivery, ethnicity, gestational age, maternal infection, and antibiotic use appear to be influential (13–16). Potential mechanisms by which bacteria enter human milk include translocation of bacteria from the maternal GI tract to the mammary gland and retrograde inoculation of bacteria from the infant's oral cavity via breastfeeding (17). Factors that influence either of these pathways, such as maternal diet and direct breastfeeding versus bottle feeding, have the potential to alter the human milk microbiota.
Diet has been consistently shown to alter the adult human GI microbiota, with high-fiber and high-fat diets both being strong predictors of microbial composition (18, 19). To our knowledge, only 2 studies have examined the association between maternal diet and the milk microbiota (20, 21); however, both studies only included metabolically healthy participants and, as a result, it is unclear whether the relation between maternal diet and the milk microbiota differs in women with metabolic dysfunction, which is known to be associated with different GI microbiota compositions (22). Furthermore, neither study examined functional capabilities of the milk microbiota, nor did they control for key confounding factors (e.g., maternal BMI) that are known to be associated with the milk microbiota. Additionally, limited data exist on how infant feeding practices (e.g., exclusive human milk vs. mixed formula feeding, frequency of direct breastfeeding per day) are associated with the human milk microbiota.
We have previously examined the association between BMI, glucose tolerance status, mode of delivery, and ethnicity with the human milk microbiota at 3 mo postpartum in a cohort of women with high rates of gestational glucose intolerance (23). The objective of this current study is to extend this initial work and investigate whether maternal diet and infant feeding practices are associated with the milk microbiota in these women.
Methods
Study design and participants
This was a cross-sectional exploratory analysis within a previously conducted prospective cohort study (ClinicalTrials.gov identifier: NCT01405547). A detailed study protocol of the original study has been previously published, as have details on the macronutrient, insulin, and adiponectin composition of milk samples, along with the association between diet and glucose tolerance status (24–26). Pregnant women were recruited between March 2009 and July 2010 from outpatient clinics at Mount Sinai Hospital in Toronto, Canada, a tertiary care center, which follows higher-risk pregnancies. Mothers provided a milk sample at 3 mo postpartum. Inclusion criteria for the original study included pregnant women ≥20 y of age with an intention to breastfeed; the current study also required an available milk sample and a complete 3-mo postpartum food frequency questionnaire (FFQ). Exclusion criteria included pre-existing diabetes diagnosis and current use of insulin (26). Written informed consent was obtained from all women and the study protocol was approved by the Sinai Health Human Research Ethics Board (24).
Collection of maternal demographic, clinical, and anthropometric data
The first study visit occurred in late pregnancy (30 wk; 95% CI: 25, 33 wk) at which time demographic and anthropometric data were collected (e.g., age, ethnicity, weight, height), and mothers provided their prepregnancy weight. All participating women underwent a 3-h 100-g oral glucose tolerance test (OGTT) during their first study visit to determine glucose tolerance status. Details of the OGTT have been published previously (25). A diagnosis of gestational diabetes, impaired glucose tolerance, or normoglycemia was made based on the results of the OGTT.
Maternal dietary assessment
Maternal dietary intake was collected via a modified Block FFQ designed to capture dietary intake from delivery to 3 mo postpartum (25, 27, 28). A paper copy of this FFQ was self-administered at the same study visit when mothers provided a milk sample. The Block FFQ has been validated against 24-h food recalls in a number of populations, including pregnant women and women living in Ontario, Canada (29–32). Detailed information regarding the FFQ and how it was administered have been published previously (25). Following completion, the FFQs were analyzed by Block Dietary Data Systems using the USDA Food and Nutrient database (33). Dietary intake values were considered outliers if they were beyond the lower [quartile 1 −1.5 × IQR (inter-quartile range)] or upper bound limits (quartile 3 +1.5 × IQR) and were deemed clinically implausible. Using this approach, a total of 6 dietary intake values were considered outliers (34–36). This resulted in 3 subjects being removed from our analyses due to ≥1 of their dietary intake values being classified as outliers.
Human milk sample collection
At the 3-mo postpartum study visit, mothers expressed breast milk using a sanitized double electric breast pump, a sterile pumping kit (e.g., breast shields, membranes, tubing; Medela, Inc.) and collection tubes. Mothers were asked to carry out a complete breast expression, if possible, and to not express milk via breastfeeding or pumping for 2 h prior to the study visit. Milk sampling was not conducted aseptically, as mothers did not wash their breasts beforehand; however, they were instructed to wash their hands prior to milk expression. Any study personnel who assisted the mother with milk expression or in handling samples wore gloves. Following milk expression, whole human milk samples were divided into aliquots and stored at −80°C until the time of microbial analyses. Infant feeding characteristics were also collected at 3 mo postpartum; a questionnaire was administered to mothers, which asked if infants were receiving exclusive human milk, how many times the infant was fed at the breast per day versus pumped milk in a bottle, and if the infant received formula, noting both frequency and quantity per day.
DNA extraction, amplification, and sequencing of human milk samples
Aliquots of whole human milk (1 mL) were pelleted by centrifugation (10,000 × g; 10 min; −80°C) and the fat layer as well as the majority of supernatant were removed. Approximately 100 μL of supernatant was used to resuspend the pellet for downstream DNA extraction. Metagenomic DNA was extracted using the NucleoSpin Food DNA Isolation Kit (Macherey-Nagel) at the Centre for the Analysis of Genome Evolution and Function at the University of Toronto, according to the manufacturer's instructions with modifications as we described previously (37). A reduced volume of elution buffer was used (30 μL) instead of the recommended 100 μL, to ensure an adequate DNA yield.
The 515F and 806R primers were used during PCR to amplify the V4 hypervariable region (38, 39). Details of PCR reactions have been described previously (37). Twenty-eight cycles of PCR were run to amplify (in triplicate) the V4 hypervariable region of the 16S ribosomal RNA (rRNA) gene. All resulting amplicons were run on a 1% Tris-Borate electrophoresis buffer agarose gel to ensure accuracy (amplicon size ∼390 bp). A negative control (sterile water), positive control (Pseudomonas aeruginosa), and mock community were also run through DNA extraction, PCR, and sequencing steps to verify accuracy. Bands of the same size and intensity were combined to generate the sequence library. The sequencing library was then purified using AMPure XP beads (0.8× volume of beads to 1× volume of library DNA) following the manufacturer's protocol and the Qubit High Sensitivity DNA Kit (ThermoFisher Scientific) was used to quantify the purified library. This finalized library was then loaded on an Illumina MiSeq and sequenced using the MiSeq-V2-300 cycle chemistry to generate 150-bp paired end reads.
Bioinformatic analyses
The raw paired end sequences from the MiSeq instrument have been deposited to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA516669. The UPARSE pipeline (USEARCH) was used for sequence analysis. The Ribosomal Database Project 16S gold database (USEARCH) was used to detect chimeric sequences in the operational taxonomic units (OTUs) and final sequences were clustered into OTUs at 97% similarity as we and others have previously described (37, 40).
Data analysis and statistics
R (version 3.4.1) and the package “Phyloseq” (version 1.27.6) were used to analyze microbiota composition (41). Singleton and doubleton OTUs were removed (i.e., OTUs that only appeared once or twice across all samples) and samples were rarefied to 20,000 reads per sample prior to downstream analyses.
ɑ-Diversity indices (Chao1/Shannon indices) were determined using Phyloseq, and associations between maternal nutrition and infant feeding characteristics with ɑ-diversity metrics as outcomes were ascertained in SAS version 9.4 (SAS Institute) using multivariable linear regression models (PROC MIXED). Two separate multivariable models were run for each outcome due to sample size constraints. Model 1 summarized overall dietary parameters and infant feeding practices, while model 2 focused on subtypes of fat and fiber. The independent variables included in model 1 were as follows: energy (kilocalories/day), total fat (percentage of energy/day), total fiber (grams/day), exclusivity of human milk (vs. mixed feeds with formula), frequency of direct breastfeeding per day (times fed at the breast, n/day), complete breast expression (yes/no), pre-pregnancy BMI (kg/m2), DNA extraction batch, and PCR sequencing batch. The independent variables included in model 2 were as follows: saturated fat (grams/day), trans fat (grams/day), polyunsaturated fat (grams/day), monounsaturated fat (grams/day), fiber from grains (grams/day), pre-pregnancy BMI (kg/m2), complete breast expression (yes/no), DNA extraction batch, and PCR sequencing batch. Multicollinearity was assessed between independent variables prior to building each model, using a variance inflation cutoff of >5. Percentage of carbohydrate was found to be collinear with total fiber and was therefore excluded from model 1. Similarly, neither percentages of protein nor fiber from fruits and vegetables were found to be associated with outcomes in any model and were therefore removed from the analyses to allow for adjustment of pre-pregnancy BMI, which we previously identified as a strong predictor of human milk microbiota composition (23). Significance was set at P < 0.05 for all ɑ-diversity results.
Phyloseq was used to ascertain B-diversities (weighted UniFrac distances and Bray-Curtis dissimilarity) and create principal coordinate analysis (PCoA) plots. Associations between maternal nutrition, infant feeding practices, and B-diversity were examined using the Adonis function in the R package “vegan” (version 2.5.4) (42). Adonis measures the amount of variation in B-diversity that is explained by each maternal or infant clinical variable. Unadjusted and adjusted Adonis models [which included pre-pregnancy BMI, complete breast expression (yes/no), DNA extraction batch, and PCR sequencing batch] were run separately with each maternal diet and infant feeding practice variable. Models including human milk exclusivity were additionally adjusted for the number of times fed at the breast, and vice versa. Since Adonis models can only include categorical variables, all continuous independent variables were categorized into quartiles for B-diversity analyses.
Differentially abundant taxa based on maternal nutrition and infant feeding practices were assessed by means of a multivariable Poisson regression model (PROC GENMOD) in SAS version 9.4 (SAS Institute). The same independent variables as described previously for ɑ-diversity (models 1 and 2) were used to assess the association between maternal nutrition and infant feeding characteristics with the relative abundance of the top 5 phyla and top 10 genera as outcome measures. To account for overdispersion in the data, the SEs of these models were adjusted using Pearson scaling. A Benjamini-Yekutieli cut-point approach was also used to account for multiple testing; P ≤ 0.022 at the phylum level (5 tests) and P ≤ 0.017 (10 tests) at the genus level were considered statistically significant for the overall group effect.
Predicted functional capabilities of human milk microbiota
Functional capabilities of the microbiota present in human milk samples were estimated using the metagenomic inference tool, Piphillin (43). The results were matched to the KEGG (Kyoto Encyclopedia of Genes and Genomes) reference database to predict the KEGG orthologs and pathways present based on 16S rRNA gene data. KEGG pathways not expressed in bacteria were subsequently removed. “MaAsLin2” (version 0.99.12) in R was used to determine statistically significant associations (P < 0.10) between maternal nutrition and infant feeding characteristics and differentially expressed KEGG pathways. The B-diversity of predicted KEGG orthologs was also assessed (Bray-Curtis dissimilarity index) using Adonis models to explore how maternal diet and infant feeding practices are associated with the overall composition of predicted microbial functions in human milk. These Adonis models were similarly adjusted for prepregnancy BMI, complete breast expression (yes/no), DNA extraction batch, and PCR sequencing batch.
Results
Participant description
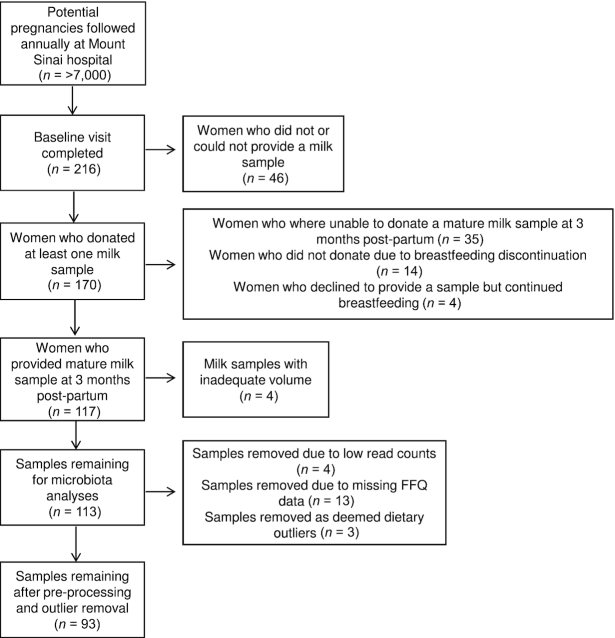
Of the 216 pregnant women who completed a baseline visit for the original study, 117 women donated a mature milk sample, with 113 samples available for this study (Figure 1). After preprocessing of the data and removal of missing data and outliers, a total of 93 mothers were included in this present analysis. Complete maternal demographic, metabolic, and obstetrical data for the cohort, including pre-pregnancy BMI, glucose-tolerance status, mode of delivery, and ethnicity are described in Table 1. On average, human milk samples were expressed and collected at 3 (SD: 1) mo postpartum and 53 (57.0%) of those samples were from a complete breast expression. At the time of milk collection, 47 (50.5%) mothers were providing their infants exclusively with human milk, compared with 46 (49.5%) mothers who provided their infants with mixed feeds of human milk and formula. On average, infants were fed at the breast 6.9 (2.4) times/d.
FIGURE 1.
Flow diagram of mothers included in this study. FFQ, food-frequency questionnaire.
TABLE 1.
Summary of maternal demographic, metabolic, and obstetric characteristics and infant feeding practices1
| Characteristic | Values |
|---|---|
| Baseline, gestation, and delivery | |
| Ethnicity | |
| White | 53 (57.0) |
| Asian (Chinese, Korean, Japanese, Filipino) | 21 (22.6) |
| Other (South Asian, black, other) | 19 (20.4) |
| Prepregnancy BMI, kg/m2 | 24.6 ± 4.6 |
| Healthy (18.5–24.9) | 57 (61.3) |
| Overweight (25–29.9) | 27 (29.0) |
| Obese (>30) | 9 (9.7) |
| Glucose tolerance status | |
| Gestational diabetes mellitus | 21 (22.6) |
| Impaired glucose tolerance | 16 (17.2) |
| Normoglycemic | 56 (60.2) |
| Mode of delivery | |
| Vaginal | 52 (55.9) |
| Scheduled cesarean section | 19 (20.4) |
| Unscheduled cesarean section | 22 (23.7) |
| Three months postpartum | |
| Current BMI, kg/m2 | 26.5 ± 5.0 |
| Healthy (18.5–24.9) | 38 (40.9) |
| Overweight (25–29.9) | 41 (44.1) |
| Obese (>30) | 14 (15.0) |
| Human milk exclusivity | 47 (50.5) |
| Mixed formula feeding | 46 (49.5) |
| Frequency of direct breastfeeding, n/d | 6.9 ± 2.4 |
Values are means ± SDs or frequencies (%); n = 93.
Mothers consumed a mean energy intake of 1776 (SD: 470) kcal/d during the first 3 mo postpartum, with 36.2% (5.0%) of those calories as fat, 16.2% (2.9%) as protein, and 49.0% (5.9%) as carbohydrates (Table 2). On average, total fiber intake was 18.5 (6.4) g/d, with a fairly equal distribution of fiber derived from both fruits/vegetables [7.8 (3.7) g/d] and grains [8.5 (4.2) g/d].
TABLE 2.
Daily nutrient intakes by women from delivery to 3 mo postpartum collected via a validated food-frequency questionnaire1
| Nutrient intakes | Values |
|---|---|
| Energy, kcal/d | 1776 ± 470 |
| Dietary fat, % of energy/d | 36.2 ± 5.0 |
| Saturated fat, g/d | 22.4 ± 7.2 |
| Polyunsaturated fat, g/d | 15.3 ± 5.3 |
| Monounsaturated fat, g/d | 28.5 ± 8.9 |
| Trans fat, g/d | 2.1 ± 0.8 |
| Dietary protein, % of energy/d | 16.2 ± 2.9 |
| Dietary carbohydrate, % of energy/d | 49.0 ± 5.9 |
| Total fiber, g/d | 18.5 ± 6.4 |
| Fiber from fruits and vegetables | 7.8 ± 3.7 |
| Fiber from grains | 8.5 ± 4.2 |
Values are means ± SDs, n = 93.
The relative abundance of microbiota in human milk has been previously described for this cohort (23); the predominant taxa in human milk samples at the phylum and genus levels are shown in Supplemental Figures 1 and 2, respectively.
Relations between maternal dietary intake, infant feeding practices, and microbiota diversity in human milk
ɑ-Diversity (within-sample diversity) was assessed using Chao1 and Shannon indices to examine the microbial richness and microbial diversity, respectively, in each human milk sample (Table 3, Supplemental Figures 3 and 4). No statistically significant associations were found between maternal nutrition, infant feeding practices, and microbial richness (Chao1 index); however, intake of both polyunsaturated fat [β estimate (SE): 0.036 (0.018); P = 0.047] and fiber from grains [0.027 (0.013), P = 0.048] were positively associated with microbial diversity (Shannon index) in multivariable linear regression analyses.
TABLE 3.
Associations between maternal diet, infant feeding practices, and ɑ-diversity of the milk microbiota1
| Chao1 index | Shannon index | |
|---|---|---|
| Model 1: overall maternal dietary intakes and infant feeding practices | ||
| Energy, kcal/d | 0.0033 (0.044) | −0.00012 (0.00013) |
| Dietary fat, % of energy/d | 0.41 (3.63) | 0.014 (0.011) |
| Total fiber, g/d | 2.47 (3.24) | 0.017 (0.0097) |
| Frequency of direct breastfeeding, n/d | 2.14 (7.80) | −0.01 (0.02) |
| Mixed feeds vs. human milk exclusivity | −3.85 (38.36) | −0.085 (0.11) |
| Model 2: subtypes of fat and fiber, g/d | ||
| Saturated fat | 3.29 (4.61) | −0.014 (0.014) |
| Polyunsaturated fat | 8.51 (5.99) | 0.036 (0.018)* |
| Monounsaturated fat | −3.03 (3.65) | −0.0081 (0.011) |
| Trans fat | −52.55 (38.42) | −0.028 (0.11) |
| Fiber from grains | 5.46 (4.50) | 0.027 (0.013)* |
Values are β estimates (SEs), n = 93. *P < 0.05. Associations were assessed via multivariable linear regression models (PROC MIXED; SAS Institute).
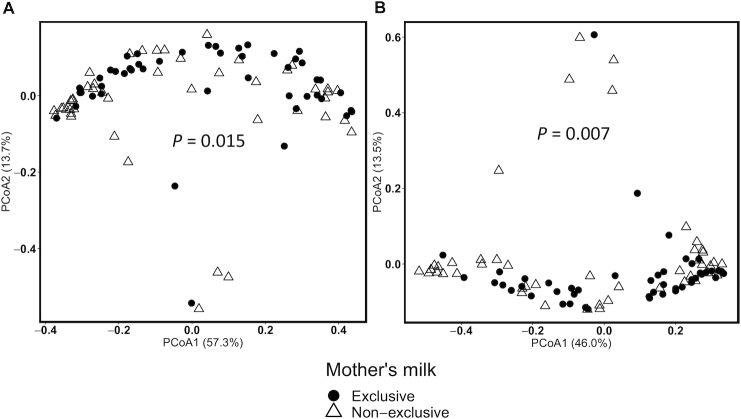
B-Diversity (between-sample diversity) was used to explore the relations between maternal nutrition and infant feeding practices with the overall microbial composition of human milk samples (Table 4, Figure 2). Human milk microbiota clustered according to human milk exclusivity (vs. mixed feeds with formula), even after adjustment for pre-pregnancy BMI, DNA extraction batch, PCR sequencing batch, and complete breast expression (weighted UniFrac R2 = 0.034, P = 0.015; Bray-Curtis R2 = 0.041, P = 0.007). Similarly, fiber from grains (R2 = 0.055, P = 0.040) and the frequency of direct breastfeeding per day (R2 = 0.057, P = 0.026) were also associated with B-diversity when applying the Bray-Curtis dissimilarity index.
TABLE 4.
Associations between maternal diet, infant feeding practices, and the B-diversity of the milk microbiota1
| Weighted UniFrac distance | Bray-Curtis dissimilarity | |||
|---|---|---|---|---|
| Characteristics | Unadjusted | Adjusted | Unadjusted | Adjusted |
| Overall dietary intakes and infant feeding practices | ||||
| Energy, kcal/d | 0.020 | 0.015 | 0.022 | 0.022 |
| Dietary fat, % of energy/d | 0.014 | 0.014 | 0.021 | 0.019 |
| Total fiber, g/d | 0.046 | 0.044 | 0.054 | 0.046 |
| Frequency of direct breastfeeding, n/d | 0.047 | 0.053 | 0.054 | 0.057* |
| Mixed feeds vs. human milk exclusivity | 0.025 | 0.034* | 0.030* | 0.041** |
| Subtypes of fat and fiber, g/d | ||||
| Saturated fat | 0.021 | 0.015 | 0.026 | 0.023 |
| Polyunsaturated fat | 0.023 | 0.023 | 0.024 | 0.023 |
| Monounsaturated fat | 0.015 | 0.012 | 0.022 | 0.020 |
| Trans fat | 0.021 | 0.015 | 0.025 | 0.023 |
| Fiber from grains | 0.033 | 0.041 | 0.049 | 0.055* |
Values are R2, n = 93. Except for mixed feeds vs. human milk exclusivity, variables were expressed as quartiles in the analysis. Associations were assessed via Adonis models. *P < 0.05; **P < 0.01.
FIGURE 2.
Representative PCoA plots of B-diversity of the human milk microbiota (n = 93). Weighted UniFrac distance (A) and Bray-Curtis dissimilarity (B) B-diversity metrics are shown. Circles denote mothers who exclusively breastfed and triangles denote mothers who provided mixed feeds. P values from multivariable Adonis analyses are shown in each panel. PCoA, principal coordinates analysis.
Relations between maternal dietary intake, infant feeding practices, and abundance of predominant taxa in human milk
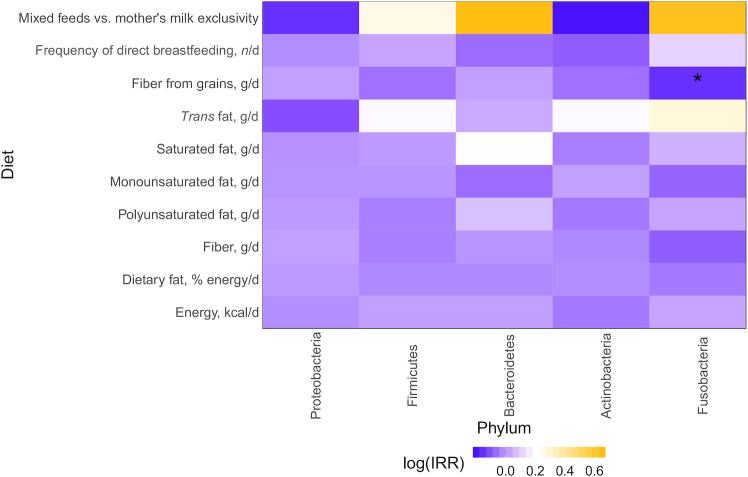
Of the top 5 phyla, only Fusobacteria showed associations with maternal diet and infant feeding practices, such that every 1-g increase in fiber from grains was associated with a reduced incidence of Fusobacteria [incidence rate ratio (IRR): 0.86; 95% CI: 0.77, 0.97; Figure 3, Supplemental Table 1].
FIGURE 3.
Differential abundance of the 5 most abundant microbiota phyla in human milk at 3 mo postpartum based on maternal diet and infant feeding practices. Heatmaps represent the results from multivariable Poisson regression models (PROC GENMOD; SAS Institute) with the top 5 phyla as the outcome variables; the colored boxes refer to the IRR on a log scale, whereas asterisks refer to statistically significant results (*P ≤ 0.022). IRR, incidence rate ratio.
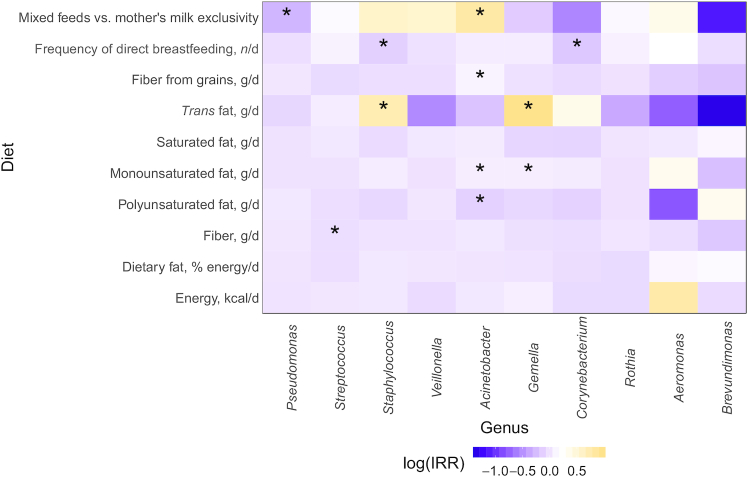
There were, however, a number of genus-level associations with maternal diet, particularly for fiber and fat (Figure 4). Each 1-g increment of fiber intake from grains was associated with an increased incidence of Acinetobacter (IRR: 1.10; 95% CI: 1.04, 1.16) and every 1-g increase in total fiber was associated with a reduced incidence of Streptococcus (0.96; 0.92, 0.99). In terms of maternal fat intake, trans fats showed a positive association with both Staphylococcus (2.07; 1.18, 3.62) and Gemella (2.63; 1.38, 5.02) incidence. Similarly, every 1-g increase in monounsaturated fat intake was associated with an increased incidence in both Acinetobacter (1.07; 1.03, 1.11) and Gemella (1.07; 1.01, 1.14). However, negative associations were observed between polyunsaturated fat intake and Acinetobacter incidence (0.89; 0.82, 0.96). No associations were observed with energy, total dietary fat, or saturated fat and the predominant genera in human milk.
FIGURE 4.
Differential abundance of the 10 most abundant microbiota genera in human milk at 3 mo postpartum based on maternal diet and infant feeding practices. Heatmaps represent the results from multivariable Poisson regression models (PROC GENMOD; SAS Institute) with the top 10 genera as the outcome variables; the colored boxes refer to the IRR on a log scale, whereas asterisks refer to statistically significant results (*P ≤ 0.017). IRR, incidence rate ratio.
When examining how infant feeding practices were associated with the human milk microbiota, it was found that mothers who fed their infants exclusively human milk (vs. mixed feeds with formula) had a reduced incidence of Pseudomonas (IRR: 0.75; 95% CI: 0.60, 0.95) and an increased incidence of Acinetobacter (2.24; 1.28, 3.96) in their milk (Figure 3, Supplemental Table 1). Furthermore, every additional time an infant was fed at the breast per day was associated with reduced incidences of Corynebacterium (0.84; 0.75, 0.94) and Staphylococcus (0.88; 0.80, 0.97).
Relations between maternal nutrition, infant feeding practices, and predicted functional capabilities of the milk microbiota
No statistically significant associations were found with maternal nutrition or infant feeding practices and predicted KEGG pathways of the milk microbiota. However, differences in the B-diversity of the overall composition of KEGG orthologs were observed with total fiber consumption (Bray-Curtis R2 = 0.067, P = 0.036) and human milk exclusivity versus mixed feeds with formula (Bray-Curtis R2 = 0.041, P = 0.013) in adjusted models (Supplemental Table 2).
Discussion
Metabolic dysfunction is known to be associated with an individual's GI microbiota and dietary patterns; however, our results are the first to provide evidence that maternal diet and infant feeding practices are also associated with the human milk microbiota at 3 mo postpartum in women with varying degrees of gestational glucose intolerance. More specifically, changes in overall microbial communities, predicted microbial functions, and individual taxa in human milk were associated with maternal dietary fiber and fat intake, as well as infant feeding practices including human milk exclusivity and frequency of direct breastfeeding per day.
Maternal consumption of fiber, particularly from grains, was associated with small but statistically significant changes in microbial community structure (ɑ- and B-diversity) and overall predicted functional profiles (B-diversity of KEGG orthologs), in addition to negative associations with Fusobacteria and Streptococcus, and positive associations with Acinetobacter abundances. To our knowledge, only 2 studies have examined relations between maternal nutrition and the milk microbiota (20, 21), one of which found positive correlations between insoluble fiber intake and the phylum Actinobacteria (primarily from the genus, Rothia) and negative correlations between soluble fiber intake and the Firmicutes phylum (20). The previously observed negative association between fiber intake and Firmicutes (20), and our observation with Streptococcus (a member of the Firmicutes phylum), is intriguing. While many Firmicutes are known to ferment complex carbohydrates to produce short chain fatty acids (SCFAs) that are beneficial to the host, such as butyrate, Streptococcus is not believed to be a major butyrate producer (44). It is important to note, however, that the use of 16S rRNA gene sequencing provides relative rather than absolute quantification of bacteria; as such, it is possible that maternal consumption of fiber alters the absolute abundance of milk microbes (e.g., members of the Firmicutes phylum) but not necessarily their relative abundance. Future work with more detailed accounting of dietary fiber sources (e.g., using weighted 3-d food records), shotgun metagenomic sequencing to enable strain-level resolution of microbiota, and absolute measures of bacteria (e.g., qPCR) would provide a more thorough understanding of these relations.
Similar to fiber, maternal consumption of dietary fat was also associated with the milk microbiota at 3 mo postpartum. Intake of polyunsaturated fat was associated with increased microbial diversity (Shannon index) and lower Acinetobacter abundance; however, the consumption of trans fat was positively associated with Staphylococcus and Gemella abundance, similar to the positive associations with monounsaturated fat intake with Acinetobacter and Gemella abundances. Previously, Williams and colleagues (20) (21 healthy mothers, 104 milk samples; United States) found maternal saturated and monounsaturated fat intake to be associated with Corynebacterium abundance in human milk over the first 6 mo postpartum, while Padilha and colleagues (21) (94 healthy mothers; Brazil) found no relations between maternal fat intake and the milk microbiota during the first month postpartum. Inconsistencies in these findings may arise from methodological differences in dietary assessment tools (e.g., FFQ vs. 24-h dietary recall, period captured during pregnancy and/or lactation), sample preparation (e.g., extraction kits, hypervariable regions amplified), and statistical analyses (e.g., adjustment for confounders vs. unadjusted correlations, corrections for multiple comparisons); however, biological differences may also explain these differences including cohort demographics and variation in geographic location, which are known to shape the microbiota of human milk (15). Future studies are needed to explore the interactions between maternal diet, human milk microbiota, and the microbial colonization and health of infants.
In addition to maternal nutrition, infant feeding practices were also associated with the microbial composition and predicted function of human milk. Exclusive human milk feeding (vs. mixed formula feeding) was associated with altered overall microbial composition (B-diversity), overall predicted microbial functions (B-diversity of KEGG orthologs), lower Pseudomonas abundance, and increased Acinetobacter abundance in human milk at 3 mo postpartum. Interestingly, Moossavi and colleagues (45) (393 healthy mother-infant dyads; Canada) recently showed higher abundances of Pseudomonas in human milk at 3 mo postpartum among mothers who pumped their milk. Similarly, mothers in our cohort who provided mixed feeds were also more likely to pump their milk versus directly breastfeed. These complementary findings provide further evidence that pumping milk is associated with an increased abundance of Pseudomonas in human milk, possibly derived from the breast pump microenvironment paired with the lack of contact with the infant oral cavity. Additionally, the provision of formula has the potential to alter the infant's oral microbiota and contribute to the microbial communities found in human milk via a backwash mechanism (17), further explaining the differences observed with exclusive human milk versus mixed formula feeding.
Similarly, increasing the frequency of direct breastfeeding per day was associated with changes in the overall microbial composition of human milk and reduced abundances of Corynebacterium and Staphylococcus. Biagi and colleagues (46) (16 mothers of preterm infants, 68 milk samples; Italy) similarly found that latching and direct breastfeeding had a significant impact on the B-diversity of both the infant oral microbiota and human milk microbiota. Additionally, both Corynebacterium and Staphylococcus are common skin-associated microbes and are known to populate the maternal areolar skin (7). Although their reduced abundance with increased direct breastfeeding frequency seems counterintuitive, it is possible that repeated exposure of the breast skin to the infant's oral cavity washes away some of these microbes, thereby diluting the abundance of skin-associated bacteria at the expense of other oral-associated microbes in human milk. As previously mentioned, the microbial changes observed in this study are relative and not absolute abundances; therefore, future studies exploring absolute concentrations of these bacteria, both on the human areolar skin and in mother's milk, in response to direct breastfeeding frequency are needed.
Strengths of the current study include its large unique cohort of women with high rates of gestational glucose intolerance, use of a validated FFQ, prediction of microbial functions to corroborate taxonomic-based microbial results, and statistical adjustment for known confounders (e.g., maternal BMI). A potential limitation of the current study is that milk was not collected aseptically, and thus, milk samples included skin microbiota and breast pump contaminants. Further, it is possible that this contamination may have masked additional diet–microbiota associations in our human milk samples; however, this collection method and subsequent data are more clinically relevant as mothers do not routinely disinfect their breast prior to pumping or direct breastfeeding. In addition, our cross-sectional analytic study design of the current analysis did not allow us to establish cause and effect between maternal nutrition, infant feeding practices, and the mother's milk microbiota, or investigate change over the postpartum period.
In conclusion, this large cohort of mothers with high rates of gestational glucose intolerance demonstrated that dietary factors, including maternal consumption of fat and fiber, and infant feeding practices, such as human milk exclusivity and frequency of direct breastfeeding, were important determinants of the human milk microbiota at 3 mo postpartum. Given the potential for microbiota in human milk to colonize the infant's GI tract, future research is needed to explore the implications of these findings on infant microbial colonization and health.
Supplementary Material
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows—SHL, AJH, and DLO: designed the research; LL-N, SHL, JKC, and PWW: conducted the research; LL-N, AK, JKC, and MRA: analyzed data and were involved in performing statistical analyses; LL-N: wrote the initial draft of the manuscript, with strong contributions from MRA and JB; all authors: participated in writing and critical revision, including SU and AS, of the manuscript for important intellectual content; DLO: had primary responsibility for final content; and all authors: read and approved the final manuscript.
Notes
Funding was provided by the Canadian Institutes of Health Research (MOP#125997) and a Canadian Diabetes Association Operating Grant (#OG-3-09-2393). SHL was supported by grant P20GM109036 from the National Institute of General Medical Sciences of the National Institutes of Health. LL-N is a recipient of a Canadian Institutes of Health Research Doctoral Award (The Frederick Banting and Charles Best Canada Graduate Scholarship Doctoral Award).
Author disclosures: AS is a co-founder of MedBiome, a clinical microbiomics company. DLO is a member of the Journal of Nutrition’s Editorial Board. The other authors report no conflicts of interest.
The sources of support had no role in the research study design or conduct, sample preparation, data or statistical analysis, interpretation, or writing of the manuscript.
Supplemental Tables 1 and 2 and Supplemental Figures 1–4 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at http://jn. nutrition.org.
Abbreviations used: FFQ, food frequency questionnaire; GI, gastrointestinal; IRR, incidence rate ratio; KEGG, Kyoto Encyclopedia of Genes and Genomes; OGTT, oral glucose tolerance test; OTU, operational taxonomic unit; rRNA, ribosomal RNA.
Contributor Information
Lauren LeMay-Nedjelski, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada; Translational Medicine Program, The Hospital for Sick Children, Toronto, Ontario, Canada.
Michelle R Asbury, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada; Translational Medicine Program, The Hospital for Sick Children, Toronto, Ontario, Canada.
James Butcher, Ottawa Institute of Systems Biology and Department of Biochemistry, Microbiology, and Immunology, University of Ottawa, Ottawa, Ontario, Canada.
Sylvia H Ley, Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA.
Anthony J Hanley, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada.
Alex Kiss, Department of Research Design and Biostatistics, Sunnybrook Research Institute, Toronto, Ontario, Canada.
Sharon Unger, Department of Pediatrics, University of Toronto, Toronto, Ontario, Canada; Department of Pediatrics, Mount Sinai Hospital, Toronto, Ontario, Canada; Division of Neonatology, The Hospital for Sick Children, Toronto, Ontario, Canada; Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada.
Julia K Copeland, Centre for the Analysis of Genome Evolution and Function, University of Toronto, Toronto, Ontario, Canada.
Pauline W Wang, Centre for the Analysis of Genome Evolution and Function, University of Toronto, Toronto, Ontario, Canada.
Alain Stintzi, Ottawa Institute of Systems Biology and Department of Biochemistry, Microbiology, and Immunology, University of Ottawa, Ottawa, Ontario, Canada.
Deborah L O'Connor, Department of Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada; Translational Medicine Program, The Hospital for Sick Children, Toronto, Ontario, Canada; Department of Pediatrics, Mount Sinai Hospital, Toronto, Ontario, Canada; Division of Neonatology, The Hospital for Sick Children, Toronto, Ontario, Canada.
References
- 1. Boix-Amorós A, Collado MC, Mira A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front Microbiol. 2016;7:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, Rodríguez JM. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69:1–10. [DOI] [PubMed] [Google Scholar]
- 3. Heikkilä MP, Saris PEJ. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95:471–8. [DOI] [PubMed] [Google Scholar]
- 4. Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16:2891–904. [DOI] [PubMed] [Google Scholar]
- 5. Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj ALet al. . Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. Can Med Assoc J. 2013;185:385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murphy K, Curley D, O'Callaghan TF, O'Shea C-A, Dempsey EM, O'Toole PW, Ross RP, Ryan CA, Stanton C. The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci Rep. 2017;7:40597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger Ket al. . Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Angelopoulou A, Field D, Ryan CA, Stanton C, Hill C, Ross RP. The microbiology and treatment of human mastitis. Med Microbiol Immunol. 2018;207:83–94. [DOI] [PubMed] [Google Scholar]
- 9. Binns C, Lee M, Low WY. The long-term public health benefits of breastfeeding. Asia Pac J Public Health. 2016;28:7–14. [DOI] [PubMed] [Google Scholar]
- 10. Dieterich CM, Felice JP, O'Sullivan E, Rasmussen KM. Breastfeeding and health outcomes for the mother-infant dyad. Pediatr Clin North Am. 2013;60:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. WHO. Short-term effects of breastfeeding: a systematic review on the benefits of breastfeeding on diarrhoea and pneumonia mortality. [Internet] WHO. [cited 2019 Aug 21]. Available from: https://www.who.int/maternal_child_adolescent/documents/breastfeeding_short_term_effects/en/. [Google Scholar]
- 12. WHO. Long-term effects of breastfeeding: a systematic review. [Internet] WHO. [cited 2019 Aug 21]. Available from: https://www.who.int/maternal_child_adolescent/documents/breastfeeding_long_term_effects/en/. [Google Scholar]
- 13. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–51. [DOI] [PubMed] [Google Scholar]
- 14. Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. 2016;7:54–60. [DOI] [PubMed] [Google Scholar]
- 15. Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, Nicol MP, Isolauri E, Yang B, Collado MCet al. . Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. 2016;7:1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bode L, McGuire M, Rodriguez JM, Geddes DT, Hassiotou F, Hartmann PE, McGuire MK. It's alive: microbes and cells in human milk and their potential benefits to mother and infant. Adv Nutr. 2014;5:571–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moossavi S, Azad MB. Origins of human milk microbiota: new evidence and arising questions. Gut Microbes. 2019;0:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolters M, Ahrens J, Romaní-Pérez M, Watkins C, Sanz Y, Benítez-Páez A, Stanton C, Günther K. Dietary fat, the gut microbiota, and metabolic health—a systematic review conducted within the MyNewGut project. Clin Nutr. 2019;38:2504–20. [DOI] [PubMed] [Google Scholar]
- 19. Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23:705–15. [DOI] [PubMed] [Google Scholar]
- 20. Williams JE, Carrothers JM, Lackey KA, Beatty NF, York MA, Brooker SL, Shafii B, Price WJ, Settles ML, McGuire MAet al. . Human milk microbial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J Nutr. 2017;147:1739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Padilha M, Danneskiold-Samsøe NB, Brejnrod A, Hoffmann C, Cabral VP, Iaucci J de M, Sales CH, Fisberg RM, Cortez RV, Brix Set al. . The human milk microbiota is modulated by maternal diet. Microorganisms. 2019;7:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang J-Y, Kweon M-N. The gut microbiota: a key regulator of metabolic diseases. BMB Rep. 2016;49:536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LeMay-Nedjelski L, Butcher J, Ley SH, Asbury MR, Hanley AJ, Kiss A, Unger S, Copeland JK, Wang PW, Zinman Bet al. . Examining the relationship between maternal body size, gestational glucose tolerance status, mode of delivery and ethnicity on human milk microbiota at three months post-partum. BMC Microbiol. 2020;20:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ley SH, O'Connor DL, Retnakaran R, Hamilton JK, Sermer M, Zinman B, Hanley AJ. Impact of maternal metabolic abnormalities in pregnancy on human milk and subsequent infant metabolic development: methodology and design. BMC Public Health. 2010;10:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ley SH, Hanley AJ, Retnakaran R, Sermer M, Zinman B, O'Connor DL. Effect of macronutrient intake during the second trimester on glucose metabolism later in pregnancy. Am J Clin Nutr. 2011;94:1232–40. [DOI] [PubMed] [Google Scholar]
- 26. Ley SH, Hanley AJ, Sermer M, Zinman B, O'Connor DL. Associations of prenatal metabolic abnormalities with insulin and adiponectin concentrations in human milk. Am J Clin Nutr. 2012;95:867–74. [DOI] [PubMed] [Google Scholar]
- 27. Block G, Hartman AM, Dresser CM, Carroll MD, Gannon J, Gardner L. A data-based approach to diet questionnaire design and testing. Am J Epidemiol. 1986;124:453–69. [DOI] [PubMed] [Google Scholar]
- 28. Block G, Coyle LM, Hartman AM, Scoppa SM. Revision of dietary analysis software for the Health Habits and History Questionnaire. Am J Epidemiol. 1994;139:1190–6. [DOI] [PubMed] [Google Scholar]
- 29. Saldana TM, Siega-Riz AM, Adair LS. Effect of macronutrient intake on the development of glucose intolerance during pregnancy. Am J Clin Nutr. 2004;79:479–86. [DOI] [PubMed] [Google Scholar]
- 30. Block G, Thompson FE, Hartman AM, Larkin FA, Guire KE. Comparison of two dietary questionnaires validated against multiple dietary records collected during a 1-year period. J Am Diet Assoc. 1992;92:686–93. [PubMed] [Google Scholar]
- 31. Siega-Riz AM, Savitz DA, Zeisel SH, Thorp JM, Herring A. Second trimester folate status and preterm birth. Am J Obstet Gynecol. 2004;191:1851–7. [DOI] [PubMed] [Google Scholar]
- 32. Boucher B, Cotterchio M, Kreiger N, Nadalin V, Block T, Block G. Validity and reliability of the Block98 food-frequency questionnaire in a sample of Canadian women. Public Health Nutr. 2006;9:84–93. [DOI] [PubMed] [Google Scholar]
- 33. US Department of Agriculture. The USDA food and nutrient database for dietary studies, 1.0. Beltsville (MD): Agricultural Research Service, Food Surveys Research Group; 2004. [Google Scholar]
- 34. Institute of Medicine. Dietary Reference Intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein, and amino acids. [Internet] 2002; [cited 2020 May 29]. Available from: https://www.nap.edu/catalog/10490/dietary-reference-intakes-for-energy-carbohydrate-fiber-fat-fatty-acids-cholesterol-protein-and-amino-acids. [DOI] [PubMed] [Google Scholar]
- 35. Dietary Assessment Primer. [Internet]. [cited 2020 May 29]. Available from: https://dietassessmentprimer.cancer.gov/learn/outliers.html. [Google Scholar]
- 36. Rhee JJ, Sampson L, Cho E, Hughes MD, Hu FB, Willett WC. Comparison of methods to account for implausible reporting of energy intake in epidemiologic studies. Am J Epidemiol. 2015;181:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. LeMay-Nedjelski L, Copeland J, Wang PW, Butcher J, Unger S, Stintzi A, O'Connor DL. Methods and strategies to examine the human breastmilk microbiome. Methods Mol Biol. 2018;1849:63–86. [DOI] [PubMed] [Google Scholar]
- 38. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer Met al. . Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinforma. 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- 40. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McMurdie PJ, Holmes S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac Symp Biocomput. 2012;235–46. [PMC free article] [PubMed] [Google Scholar]
- 42. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos Pet al. . vegan: Community Ecology Package. [Internet] 2019; [cited 2019 Aug 21]. Available from: https://CRAN.R-project.org/package=vegan. [Google Scholar]
- 43. Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, DeSantis TZ. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11:e0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vos P, Garrity G, Jones Det al. editors. Bergey's manual of systematic bacteriology: volume 3: the Firmicutes. [Internet] 2nd ed New York: Springer-Verlag; 2009; [cited 2020 Mar 2]. Available from: https://www.springer.com/gp/book/9780387950419. [Google Scholar]
- 45. Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza RJ, Becker AB, Mandhane PJet al. . Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324–35, e4. [DOI] [PubMed] [Google Scholar]
- 46. Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, Faldella G, Candela M, Brigidi P, Corvaglia L. The bacterial ecosystem of mother's milk and infant's mouth and gut. Front Microbiol. 2017;8:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.