Abstract
Background
Pathological diagnosis of glioma subtypes is essential for treatment planning and prognosis. Standard histological diagnosis of glioma is based on postoperative hematoxylin and eosin stained slides by neuropathologists. With advancing artificial intelligence (AI), the aim of this study was to determine whether deep learning can be applied to glioma classification.
Methods
A neuropathological diagnostic platform is designed comprising a slide scanner and deep convolutional neural networks (CNNs) to classify 5 major histological subtypes of glioma to assist pathologists. The CNNs were trained and verified on over 79 990 histological patch images from 267 patients. A logical algorithm is used when molecular profiles are available.
Results
A new model of the squeeze-and-excitation block DenseNet with weighted cross-entropy (named SD-Net_WCE) is developed for the glioma classification task, which learns the recognizable features of glioma histology CNN-based independent diagnostic testing on data from 56 patients with 17 262 histological patch images demonstrated patch level accuracy of 86.5% and patient level accuracy of 87.5%. Histopathological classifications could be further amplified to integrated neuropathological diagnosis by 2 molecular markers (isocitrate dehydrogenase and 1p/19q).
Conclusion
The model is capable of solving multiple classification tasks and can satisfactorily classify glioma subtypes. The system provides a novel aid for the integrated neuropathological diagnostic workflow of glioma.
Keywords: convolutional neural networks, deep learning, glioma, histology, neuropathology
Key Points.
1. An AI neuropathological platform was developed to classify histological and molecular subtypes of glioma.
2. This model achieved promising histological diagnostic outcomes with patch-level accuracy of 86.5% and patient-level accuracy of 87.5%.
Importance of the Study.
Glioma is the most common brain tumor. Pathological diagnosis of glioma subtypes is essential for treatment plan and prognosis. However, current diagnosis heavily relies on experienced neuropathologists while personal error is almost inevitable. Therefore, we developed an “AI Neuropathologist” platform, comprising a slide scanner and deep CNNs to classify 5 major subtypes of glioma for assisting pathologists. Our model demonstrated a patch-level accuracy of 86.5% and a patient-level accuracy of 87.5%. Our CNNs learned the recognizable features of glioma histology, and can classify integrated neuropathological subtypes along with IDH and 1p/19q status. The system provides a novel aid for the neuropathological diagnostic workflow of glioma.
Gliomas are the most prevalent primary malignant brain tumors in adults, and are classified into grades I–IV based on malignant behavior by the World Health Organization (WHO).1 Until recently, histologic diagnosis by neuropathologists via microscopic visual inspection of histopathological slides has been the gold standard for classification, especially hematoxylin and eosin (H&E) sections, carrying prognostic information for patient management.2 The 2016 WHO classification of tumors of the central nervous system facilitates objective and precise analysis of glioma, not only on growth patterns and behaviors but also more pointedly on genetic markers.3
Histopathological diagnosis of glioma is a laborious process, including manually examining both coarse and fine resolutions of images covering large volumes of tissue samples. The pathologist also faces complex classification criteria, which calls for detailed and exhaustive analysis based on experience. Moreover, despite well-established grading strategies, analyses from multiple pathologists on the same sample (especially those without significantly bifurcated appearance features) can easily yield inconsistency even among experts, as they draw from different perceptions and are subject to various biases.4,5 Interobserver variability has become an issue as prognosis diversity can vary based on histological diagnosis within one subtype of glioma.6–8 The evidences shows that subjectiveness could mislead clinical decisions, and there may be a better method for analyzing the histo-features of glioma to assist human intelligence.
The standard practice of microscopic diagnosis for classification and grading of cancer has only limitedly evolved throughout the decade. In contrast to other medical examinations, pathology has not kept up with the digital revolution.9 However, in recent years, with the development of slide scanners that digitize glass slides into images, mostly known as whole-slide images (WSIs),10 advancement in scanning technologies over the past 2–3 years11 has permitted large numbers of slides to be scanned, forging the way for computational pathology and particularly the use of artificial intelligence to help pathologists with their efficiency and accuracy on diagnosis.12,13
Convolutional neural networks (CNNs) have been proven highly successful and often exceed conventional standards for a number of image analysis tasks,14 such as DenseNet and Inception-FCN.15,16 A deep learning approach to glioma classification can provide automatic and unbiased decisions to help guide and corroborate neuropathologists’ work. Ertosun et al first applied CNNs to histopathological glioma grading17; however, their model only led to binary classification of glioblastoma (GBM) and low-grade glioma (LGG), and of grades II and III among LGG samples. There will be a greater benefit from a more detailed subtype classification of gliomas, which can provide a more clinically valuable diagnosis. Thus, the immediate question is whether CNN can be applied to comprehensive glioma histopathological subtype diagnoses.
In this study, the “AI Neuropathologist for Glioma” platform is proposed, which receives high-magnification histopathological images and returns a diagnosis based on these images, together with high-speed automated image collection for pathological slides, precise patch image stitching, and diagnostic networks. The significance of the study lies in high interobserver discordance rates among neuropathologists, since gliomas have indeterminate histologic features.2 The goal is to reach the testing accuracy of at least 80% for histopathological images based on computer science methodology by using retrospective data before carrying out future clinical research. In addition, molecular markers including isocitrate dehydrogenase (IDH) and 1p/19q status are necessarily integrated into the network in accordance with the WHO 2016 classification of tumors of the central nervous system for molecular pathological diagnosis.
Materials and Methods
Digital Data Acquisition
Instead of using a manufacturer scanner with WSIs, in this study, a low-cost transformed slide scanner is developed by adding a digital camera and micropositioning platform on a routine optical microscope as shown in Fig. 1. The stained tumor section is placed on the micropositioning platform and images are captured after each movement. A total of 300 image patches of size 2048 × 1536 pixels at 400x magnification were scanned for each slide under 6 minutes, saved to our lab server, and later stitched together. The detailed methodologies can be found in Supplementary Material 1.
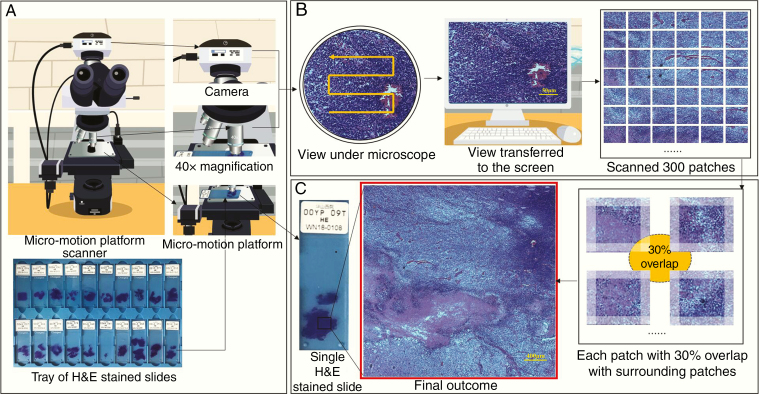
Fig. 1.
Image collection and stitching process. (A) Samples of H&E-stained slides to be placed on a micromotion platform scanner. A camera is loaded on the top of the ordinary microscope, and a fretting platform is loaded on the stage to control the motion of the slide. (B) Scanner operation mechanism: the platform moves in an S-shape as the camera takes pictures of the view under the microscope. The view is transferred to the computer screen in real time. The platform allows the glass slide to move 0.2 mm horizontally or 0.15 mm vertically each time. Images were captured under a setting of 10× eyepieces and 40× objective lenses with sizes of 2048 × 1536. As the platform captures 20 images horizontally and 15 images vertically, a total of 300 patches of images were taken and saved. (C) Image stitching: each patch (image) among 300 patches had a 30% overlap with surrounding patches, which achieved an intact pathological image as the final outcome.
A total of 97 252 histopathological images were collected from histopathological slides of 323 classified glioma patients. The dataset was obtained from the Central Nervous System Disease Biobank,18 Huashan Hospital, Fudan University, Shanghai. In this study, we adopted a patient-level split of images into 3 parts: (i) the training set included 65 673 images (from 219 subjects), (ii) the validation set included 14 317 images (from 48 subjects), and (iii) the testing set included 16 862 images (from 56 subjects). The study was ethically approved by the Huashan Hospital institutional review board. The images included 6 histological categories of glioma: oligodendroglioma (O, WHO grade II), anaplastic oligodendroglioma (AO, WHO grade III), astrocytoma (A, grade II), anaplastic astrocytoma (AA, grade III), glioblastoma (GBM, grade WHO IV), and nontumor images of red blood cells and background brain glia (BG). The detailed data distribution is shown in Fig. 2. Each pathological image was categorized into one of the above 6 subtypes by 2 trained doctors. For the cases with varied opinions, a senior pathologist reviewed the images and made the final decision. For each slide, 300 (patch) images were taken to cover most of the visible areas. Next, the image quality was checked and those with poor quality were discarded. The categorical spread of the dataset was relatively imbalanced, since the labels were given on the patch level rather than the subject level. Data were randomly split into the training and validation sets in relatively similar proportions. The training set was used to train networks. The best performing model is selected by using the validation set. Finally, an independent testing dataset of 56 patients with 17 262 histological patch images was prepared for accuracy evaluation.
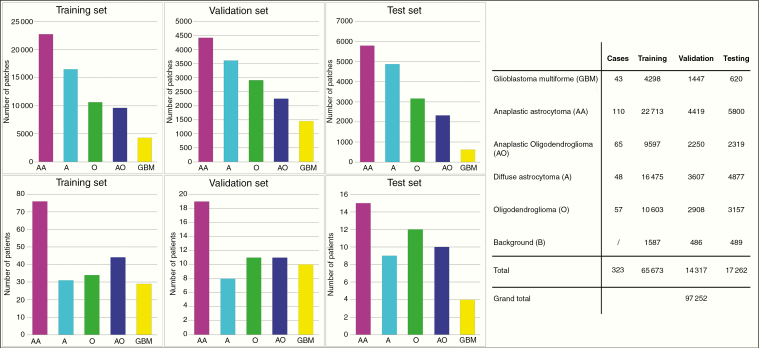
Fig. 2.
Six histological categories of glioma H&E sections were involved. 1) oligodendroglioma, 2) anaplastic oligodendroglioma, 3) astrocytoma, 4) anaplastic astrocytoma, 5) glioblastoma, and 6) nontumor images of red blood cells and background brain glia. The images were divided into 3 sets: 1) training set: 65 673 images from 219 subjects, 2) validation set: 14 317 images from 48 subjects, and 3) testing set: 16 862 images from 56 subjects. Bar charts for the number of patches and patients are displayed on the left, and detailed numbers of 5 subtypes for the training, validation and testing sets are shown on the right. Note that background image patches were taken from the boundary of tumor regions in the sections.
Diagnostic Model
The experiments were performed by using Pytorch on a Linux system and 2 NVIDIA Tesla 32G graphics processing units (GPUs). The strategy was to develop a diagnostic model based on image patches, and then combine the patch prediction results with molecular information into the final integrated neuropathological diagnosis for the patient.
All image patches were first preprocessed and resampled to size of 512 × 384 while maintaining image details and aspect ratio to save computational GPU memory. For the training stage, to improve the generation ability of our model, an image augmentation process such as random rigid transformation (including vertical/horizontal flipping and rotation) was applied to each image. To avoid inconsistency in color and luminance due to the influence of light and staining process, the brightness, contrast, and color of each training image were randomly changed before it was fed into the network.
For the image patch diagnostic model, we proposed a neural network named squeeze-and-excitation block DenseNet (SD)-Net_WCE. Briefly, we employed the DenseNet structure as the backbone, where pathology images were sent to the convolutional layers to form feature maps, and different levels of feature maps are further densely concatenated to retain the low- and high-level image features. Such a design can strengthen feature propagation and encourage feature reuse. After that, these feature maps were sent to fully convolutional layers with the softmax operator, and the output was the final prediction of glioma categories. Based on this, we added squeeze-and-excitation blocks between the batch normalization layer and rectified linear unit layer in dense layers, named SD-Net. As different cells have different boundary shapes, certain channels of feature maps generated by the corresponding convolutional kernels contributed more to the final classification task. Thus, adding SD blocks can theoretically improve the classification performance theoretically. Each SD block was followed by a transition layer, eventually connected in series to form the proposed SD-Net. Finally, a modified weighted cross-entropy19 loss was adopted as the loss function to address the issue of data imbalance between glioma categories, to replace the original cross-entropy loss function in DenseNet. SD-Net optimized by such a weighted cross-entropy loss function is named SD-Net-WCE. Additionally, to accelerate convergence, a transfer learning strategy is utilized. That is, pretraining parameters on the ImageNet dataset were used to initialize our model. The model was optimized using the Adam20 algorithm; the original learning rate was set to 10–4, and the weight decay rate was set to (0.9, 0.999). In the testing stage, the trained model was used to provide a prediction of glioma categories for a testing image patch.
Fig. 3 illustrates the image classification pipeline based on the deep learning model. To fuse image patch results at the patient level, a hierarchical strategy was employed. Despite background patches, 2 major types of oligodendroglioma (O and AO) and astrocytoma (A, AA, GBM) were distinguished based on the number of image patches. Then, based on the observation in training data, we determined that the oligodendroglioma slide would be classified into AO if its proportion (AO/(AO+O)) was greater than 10%, and into O otherwise. Similarly, the astrocytoma would be classified into GBM if its portion (GBM/(GBM+AA+A)) was greater than 10%. If not, it would be considered AA if its portion (AA/(AA+A)) was greater than 10%, and A otherwise. A detailed description of methods could be found in Supplementary Material 2.
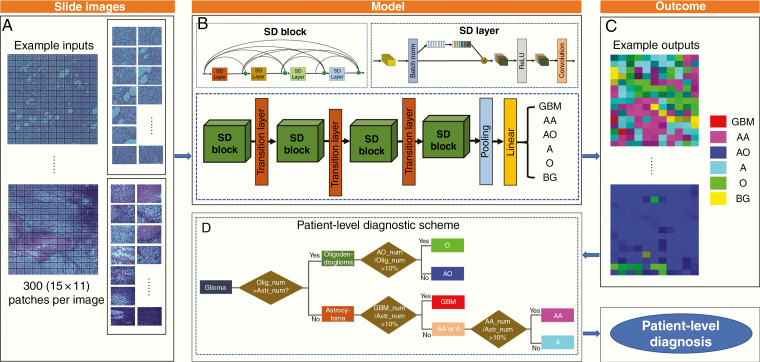
Fig. 3.
Diagram of the proposed method framework. (A) Network input: scanned slide images are sent to the CNN model in patches. (B) The whole neural network system structures of our method are provided where multiple SD blocks and transition layers are used to obtain the prediction of classifications for each patch. Subtype predictions are labeled as GBM, AA, AO, A, O, and BG. (C) Network output: visually distinguish 6 histological subtypes of glioma correspond to 6 colors, respectively. The shade of color shows different level of certainty determined by the network. (D) These patch labels go through a hierarchical decision tree for patient-level diagnosis based on the amounts and proportions of tumor types. The outcome thus includes results for both the image patch label and the patient-level label.
Integrated Neuropathological Diagnosis
The formalin-fixed paraffin-embedded tumor tissue blocks were stored in the Glioma Tissue BioBank. Tumor DNA was extracted from a representative tumor area with more than 70% tumor content. Mutational hotspots of IDH1 at codon 132 and IDH2 at codon 172 were evaluated by direct sequencing. Chromosome 1p and 19q codeletion were evaluated by fluorescence in situ hybridization.21 A detailed description of the molecular biomarker evolution process is given in the Supplementary Material 3.
When molecular pathological results were available, the above glioma categories could be extended into integrated neuropathological diagnosis. As shown in Fig. 4, we provide the integration scheme for the 2 most important molecular information carried out by the WHO 2016 classification of tumors of the central nervous system3: IDH and 1p/19q. First, each of these 5 glioma categories was split into 2 branches based on the IDH status of the given patient, similar to a decision tree. If the final classification was not reached, the 1p/19q status was used to further split the patient diagnosis into 2 branches, where the horizontal arrows lead to the final diagnosis. In this way, the molecular pathological diagnosis was obtained.
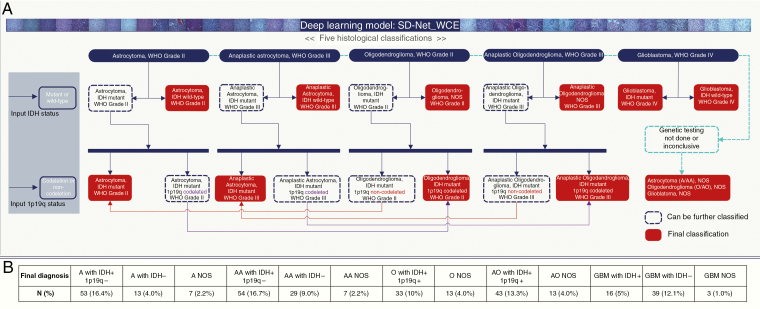
Fig. 4.
Illustration of logical algorithms. Five histological classifications with different grades of patient level prediction are demonstrated by using the deep learning model. IDH mutation and 1p/19q codeletion status are set as molecular markers. When genetic testing is not done, diagnosis conclude as “histological classification + NOS.” When molecular information of the patient is available, it could be extended into molecular pathological diagnosis by following the route of a decision tree. Final integrated classifications include A with IDH wildtype, AA with IDH wildtype, GBM with IDH mutation, GBM with IDH wildtype, A with IDH mutation and 1p/19q non-codeletion, AA with IDH mutation and 1p/19q non-codeletion, O with IDH mutation and 1p/19q codeletion, AO with IDH mutation and 1p/19q codeletion, A NOS, AA NOS, O NOS, OA NOS, and GBM with NOS. (B) Number and proportion of final diagnosis of all 323 cases after integrating IDH and 1p/19q status with histological classification in accordance with the 2016 WHO classification of tumors of the central nervous system.
Results
A CNN (model SD-Net-WCE) was developed for automated classification of pathology images into 6 subtypes. The performance of the CNN models on pathological image classification was evaluated, including both SD-Net and SD-Net-WCE. For comparison, 2 other popular methods were adopted: DenseNet and Inception-FCN. The proposed SD-Net-WCE shows the best accuracy at 87%. A detailed comparison can be found in Supplementary Material 4.
In simple terms, the overlap of each image is discarded, 165 patches are put into the model for a single case. Images pass through the SD blocks and layers to obtain 6 categories of patch-level output including GBM, AA, AO, A, O, and BG. For better visual effect, they are then assigned to different colors. Those outcomes then pass the patient-level diagnostic scheme for the final patient-level outcome. A detailed demonstration can be found in the video Hyperlinked Supplement 1—Software Operational Approach.
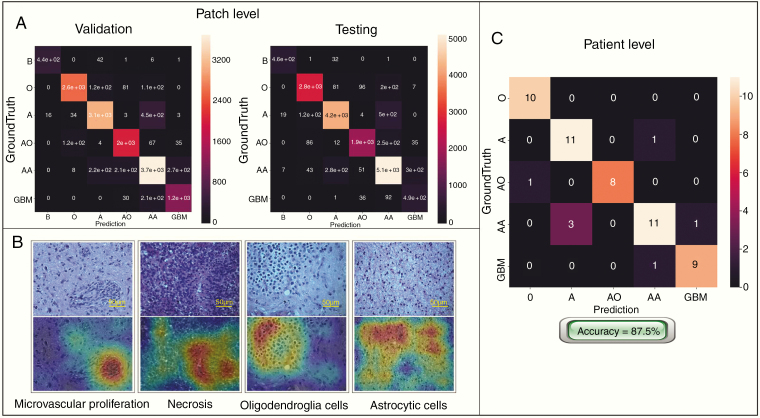
The CNNs were trained and verified on over 79 990 histological patch images from 267 patients and independently tested on data from 56 patients with 17 262 histological patch images. Table 1 shows the sensitivity and specificity across 5 classifications in 6-fold after cross-validation with an average patch accuracy of 86.5%. Fig. 5 gives the confusion matrices of the proposed SD-Net-WCE. By comparing the input testing subject, and the corresponding attention maps, the attention map can accurately highlight the tumor cell clusters, which further ensures the efficacy and robustness of our deep learning model. The image patch prediction results were then combined to complete the final classifications of the mosaic large images and produce the patient-level diagnosis. The results show that our deep CNN can learn pathological features and accomplish glioma classification tasks, since our patient-level accuracy reached 87.5%.
Table 1.
Sensitivity, specificity, and accuracy of each fold across each classification of SD-Net_WCE model
| O | A | AO | AA | GBM | Accuracy | |
|---|---|---|---|---|---|---|
| Fold 1 | 89.48 / 98.50 | 90.58 / 96.60 | 91.11 / 96.54 | 81.14 / 92.41 | 79.68 / 97.37 | 87% |
| Fold 2 | 95.10 / 96.73 | 87.10 / 96.71 | 93.57 / 95.66 | 82.09 / 96.27 | 74.21 / 95.88 | 86% |
| Fold 3 | 89.10 / 98.17 | 91.71 / 95.04 | 95.04 / 97.28 | 87.34 / 92.31 | 68.29 / 99.21 | 87% |
| Fold 4 | 87.36 / 96.87 | 87.95 / 94.21 | 85.83 / 98.43 | 84.28 / 92.70 | 68.61 / 97.79 | 85% |
| Fold 5 | 84.84 / 98.14 | 87.05 / 96.86 | 83.61 / 98.70 | 88.69 / 90.28 | 79.19 / 97.87 | 87% |
| Fold 6 | 90.39 / 98.53 | 93.22 / 97.28 | 90.51 / 97.52 | 86.91 / 95.93 | 76.81 / 97.46 | 89% |
| Average Sensitivity (%) | 89.37 | 89.60 | 89.94 | 85.08 | 74.46 | Mean accuracy: 86.5% |
| Average Specificity (%) | 97.82 | 96.12 | 97.36 | 93.32 | 97.60 |
Fig. 5.
Confusion matrices and attention maps of the SD-Net-WCE model. (A) The confusion matrix of the proposed SD-Net-WCE for the validation set and the test set, respectively. “AA” had the highest error rate and “A” were also misclassified, probably because cells of AA and A are confusing due to differences arising as a result of nuclear division—an information relatively difficult to capture. (B) Attention maps of pathological images generated by the class activation mapping (CAM) method. Images in the upper row are the pathological section images, and the lower row are the corresponding attention maps. Regions with warm color refer to the areas on which our model is focused when it separates tumor cells from other tissues. (C) Confusion matrix based on patient-level of testing group. “A” is relatively more confused with “AA.” This may be because the mitosis is small and difficult to identify as a major judgment feature for “AA.”
Ultimately, our model reached the molecular level with a total of 323 cases, including training, validation, and testing groups. During genetic testing, 7 cases had no reserved tissue blocks and 20 cases failed quality control due to the high degradation of the specimens. Those 27 cases were allocated to “not otherwise specified (NOS)” based on their histological diagnosis. Hence, 296 cases were genetically tested for IDH and 1p/19q status. All results were input into a logical algorithm derivation. Numbers of cases that fall into each final histological and molecular diagnosis are shown in Fig. 4. All data are presented in Supplementary Material 5.
Discussion
The development of pathology depends on technological advances.22 The main hypothesis addressed in this work is that the clinical-grading and classification performance of glioma can be reached by annotating at the patch level. To test our hypothesis, we developed and applied a deep learning approach to the problem of automated classification of different histopathological subtypes of glioma based on the dataset collected from Huashan Hospital using a self-developed digital slide scanner. A large dataset was compiled comprising 97 252 images from 323 slides of 323 individual cases across 5 different subtypes of glioma. We performed this through the assembly of 2 CNN components, each specialized to its tasks. Our preliminary results appear to be promising, showing 86.5% patch accuracy and 87.5% patient-level accuracy for distinguishing O, AO, A, AA, and GBM on the independent testing dataset. We take advantage of the recent advances in deep learning to train a deep neural network in the task of glioma classification. CNNs are composed of multiple layers, each of which provides an integral step in the data processing. Early in the network, the first layers can produce only low-level, basic patterns. Conversely, the deepest layers of the model can represent highly complex, abstract features that may correspond with particular classes. (See Hyperlinked Supplement 2—Animation for an overall concept of this study.
“AI Neuropathologist” is considered to be a part of translational medicine, and thus the actual clinical performance of the model is most concerning. Practically, to our knowledge, there is no applied glioma pathological diagnostic system. By looking into multiple deep learning strategies, the model we developed has given the most reliable result with comparatively high accuracy. It is also considered to be competitive compared with pathologists with low time consumption, as the automated process takes less than 10 minutes in total, including image collection, stitching, and diagnosis. Given that accurate and rapid diagnosis is the future direction of digital pathology development, our study has the potential for assisting pathologists with their diagnoses in the practical field, especially for those who lack experience in diagnosing glioma. The cell and histologic structures of different types of glioma which are considered by pathologists are also well recognized by the model. The task of classifying the type and grade of glioma using image features is not unlike other types of automated image feature learning problems, in which a set of predefined features is used to characterize the image and predict the classification label, and our work can be approached using features such as nuclear shape.23 However, a substantial disadvantage of this is the need to know those that are the most informative in the task. Often the best features are not known, and a method of unsupervised feature learning can be advantageous, particularly if abundant data are available.
Despite the general concerns about overfitting and scarcity of data, our chosen model trains well on the dataset and proves robust in validation. Although subject-level prediction will almost always yield better accuracy, the 2 methods are fairly consistent with one another and may be entirely comparable. False patch labeling, though mitigated in validation testing, remains an issue in the training phase, as the network will still learn from incorrect examples. Currently, there are no obvious solutions that can provide sufficient data augmentation without compromising the integrity of the data other than slowly gathering more samples. We found the lack of availability of high-quality histopathological images to be the greatest limiting factor in our study. The publicly available datasets from The Cancer Genome Atlas are for LGG and GBM and contain only large WSIs, each exceeding 2 GB in size. Le Hou et al approached this problem also by employing patch-based CNNs, although their model operates on data at a much lower resolution of 20x.24 Our work can benefit from a region-of-interest detection method to extract suitable image samples from WSIs and then serve as input into our current classification model.
The most recent research by Hollon et al developed a near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks.25 The basis of the study was Raman histology, which is not well popularized in the clinical histopathological field. Nevertheless, traditional histological diagnosis is still the mainstream of cancer care. Although their results showed a higher accuracy, it is unrealistic to take in this model practically at short notice considering both training and equipment expenses. In addition, the data distribution concentrated on nonglial tumors, and glioma classification by using Raman histology had a relatively poor performance among types of brain tumors, which was the evidence of requiring traditional postoperative histopathology. Deep learning models of histopathology for other disciplines are also developing rapidly these years. For example, Yu et al developed a lung cancer model to differentiate adenocarcinoma and squamocellular carcinoma.26 Similarly, other models, including skin cancer, breast cancer, prostate cancer, and cervical cancer, remain a dichotomy rather than multiclassification, which can divide images into either cancerous or noncancerous histology.10,27,28
The WHO 2016 classification of tumors of the central nervous system concludes that IDH and 1p/19q are necessary genetic features when integrated diagnosing glioma, since molecular signatures can sometimes outweigh histologic characteristics.3 Therefore, in addition to histological classification, our model has also adopted molecular diagnosis by using a logical algorithm. If molecular information of patients is available, the final diagnosis can be further amplified, as shown in Fig. 4. It now appears that the technical difficulties are the deep learning network of histological image recognition. Relatively, logical discrimination of molecular markers is clear and intelligible. Our model is capable of updating if any novel molecular marker is indicated in future WHO classification.
The result of our evaluation of accuracy for glioma classification is a reasonable preliminary result but leaves room for improvement. There are several potential reasons for this performance. First, protocols for tissue slicing, staining, image acquisition, etc, are critical. Thus, the quality of the image, specifically to patches, could take effect. The variation within the tissue slides, and portions of pathology images could contain nontumor tissue which may or may not be relevant to the diagnosis. Future work in which our methods are applied to more independent multicenter data would be good to establish the magnitude of those confounding aspects on the accuracy. Furthermore, as our model can only classify 5 major types of glioma, it is necessary to distinguish nonglial images before being allocated into glioma subtypes. These images include normal tissue and other types of brain tumors, which can be achieved by expanding both the number and kind of samples and adding another neural network in advance.
In summary, by taking advantage of deep image learning technologies, the proposed “AI Neuropathologist” can successfully classify 5 major types of histopathological glioma images and integrate with IDH and 1p/19q status according to the most recent standards outlined by the WHO 2016 classification of tumors of the central nervous system. The accuracy of the diagnostic model at both patch level and patient level has reached expectations and showed competitiveness compared with existing models. At present, the accuracy of our model has reached the general condition of carrying out prospective clinical research, and it will be the focus of the next step for our projects.
Supplementary Material
Acknowledgments
The authors thank pathologists of Department of Pathology, Huashan Hospital, Fudan University for their data labelling contribution. The authors thank Shanghai KR Pharmtech, Inc., Ltd. for genetic testing contribution.
Conflict of interest statement. Feng Shi, Qiuping Chun, and Dinggang Shen are employees in United Imaging Intelligence. Chunming Mei and Jun Zhang are employees in Zhongji Biotechnology. These two companies have no role in designing and performing the surveillances and analyzing and interpreting the data. All other authors report no conflict of interest relevant to this article.
Authorship statement. LJ, FS, JW, and DS conceived and designed the study. LJ, QC, HC, YM, SW, CM, and AA performed and analyzed the research. LJ and FS wrote the paper. NH, JL, JZ, DS, and JW reviewed and edited the manuscript. All authors read and approved the manuscript.
Funding
This project was supported by Shanghai Municipal Science and Technology Major Project (no. 2018SHZDZX01), Shanghai Brain Bank technical system (no.16JC1420103) and ZJLab.
References
- 1. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Perry A, Wesseling P. Chapter 5 - Histologic classification of gliomas. In: Berger M, Weller M, eds, Handbook of Clinical Neurology. 2016;134:71–95. [DOI] [PubMed] [Google Scholar]
- 3. Miriam N, Kurtis B, Debraj M, Manuel SJ, Black KL, Patil CG. Survival and prognostic factors of anaplastic gliomas. Neurosurgery. 2013;3(3):458–465. [DOI] [PubMed] [Google Scholar]
- 4. Van den Bent M Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuroapthologica. 2010;120(3):297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharma I, Kaur M, Mishra AK, et al. Histopathological diagnosis of leprosy type 1 reaction with emphasis on interobserver variation. Indian J Lepr. 2015;87(2):101–107. [PubMed] [Google Scholar]
- 6. Ellison DW, Kocak M, Figarella-Branger D, et al. Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed. 2011;10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108(1):49–56. [DOI] [PubMed] [Google Scholar]
- 8. Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64(19):6892–6899. [DOI] [PubMed] [Google Scholar]
- 9. Hajdu SI A note from history: microscopic contributions of pioneer pathologists. Ann Clin Lab Sci. 2011;41(2):201–206. [PubMed] [Google Scholar]
- 10. Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fuchs TJ, Buhmann JM. Computational pathology: challenges and promises for tissue analysis. Comput Med Imaging Graph. 2011;35(7-8):515–530. [DOI] [PubMed] [Google Scholar]
- 13. Louis DN, Feldman M, Carter AB, et al. Computational pathology: a path ahead. Arch Pathol Lab Med. 2016;140(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y, Gadepalli K, Norouzi M, Dahl GE. Detecting cancer metastases on Gigapixel pathology images Available at https://arxiv.org/abs/1703.02442. Accessed March 8, 2017.
- 15. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Transactions on Pattern Analysis & Machine Intelligence. 2014;39(4):640–651. [DOI] [PubMed] [Google Scholar]
- 16. Huang G, Liu Z, Laurens VDM, Weinberger KQ. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2017; Honolulu, HI; 2261–2269.
- 17. Ertosun MG, Rubin CL. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. AMIA Annual Symposium Proceedings. 2015;2015:1899–1908. [PMC free article] [PubMed] [Google Scholar]
- 18. Aibaidula A, Lu JF, Wu JS, et al. Establishment and maintenance of a standardized glioma tissue bank: Huashan experience. Cell Tissue Bank. 2015;16(2):271–281. [DOI] [PubMed] [Google Scholar]
- 19. Shore J, Johnson R. Axiomatic derivation of the principle of maximum entropy and the principle of minimum cross-entropy. IEEE Transactions on Information Theory. 2003;26(1):26–37. [Google Scholar]
- 20. Kingma DP, Ba J. Adam: a method for stochastic optimization Available at https://arxiv.org/abs/1412.6980v8. Accessed January 30, 2017.
- 21. Koh J, Cho H, Kim H, et al. IDH2 mutation in gliomas including novel mutation. Neuropathology. 2015;35(3):236–244. [DOI] [PubMed] [Google Scholar]
- 22. Bailey OT Greenfield’s neuropathology. JAMA. 1964;187(9):1001.14102934 [Google Scholar]
- 23. Das K, Karri SPK, Guha RA, Chatterjee J, Sheet D. Classifying histopathology whole-slides using fusion of decisions from deep convolutional network on a collection of random multi-views at multi-magnification. 2017 IEEE International Symposium on Biomedical Imaging (ISBI 2017). 2017; Melbourne, VIC; 1024–1027.
- 24. Le H, Samaras D, Kurc TM, Gao Y. Patch-based convolutional neural network for whole slide tissue image classification. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016; Las Vegas, NV; 2424–2433. [DOI] [PMC free article] [PubMed]
- 25. Hollon TC, Pandian B, Adapa AR, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020;26(1):52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Veta M, Pluim JP, van Diest PJ, Viergever MA. Breast cancer histopathology image analysis: a review. IEEE Trans Biomed Eng. 2014;61(5):1400–1411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.