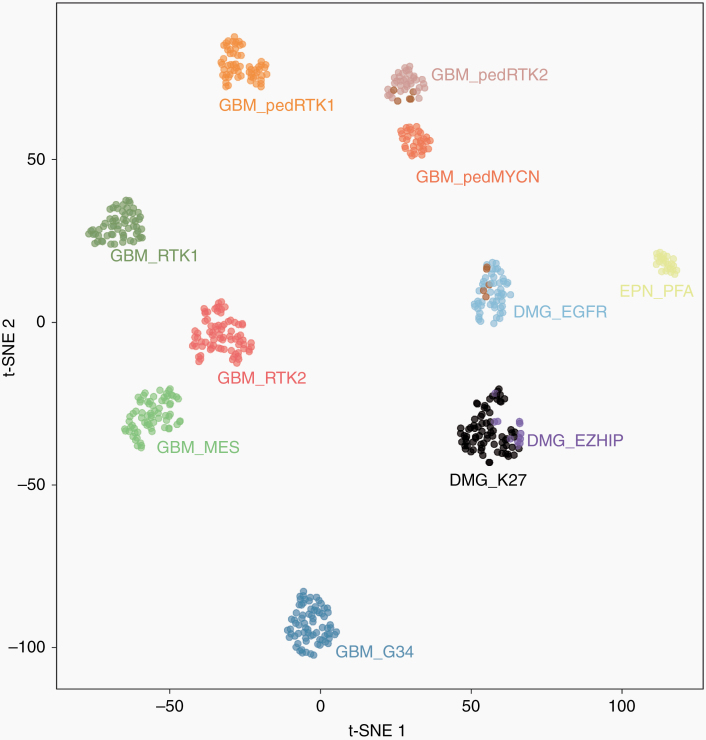
Fig. 1.
t-SNE analysis of DNA methylation profiles of the 58 gliomas investigated (DMG_EGFR; 10 of the cases included from the Mondal et al cohort (UCSF) are highlighted in brown) alongside selected reference samples. Reference DNA methylation classes: glioblastoma IDH-wildtype, subclass RTK 1 (GBM_RTK1); glioblastoma IDH-wildtype, subclass RTK 2 (GBM_RTK2); glioblastoma IDH-wildtype, subclass mesenchymal (GBM_MES); glioblastoma IDH-wildtype, pediatric RTK 1 (GBM_pedRTK1); glioblastoma IDH-wildtype, pediatric RTK 2 (GBM_pedRTK2); glioblastoma IDH-wildtype, pediatric MYCN (GBM_pedMYCN); diffuse midline glioma H3 K27M-mutant (DMG_K27M); diffuse midline glioma H3-wildtype overexpressing EZHIP (DMG_EZHIP); glioblastoma IDH-wildtype, H3.3 G34-mutant (GBM_G34); ependymoma, posterior fossa group A (EPN_PFA).