Fig. 2.
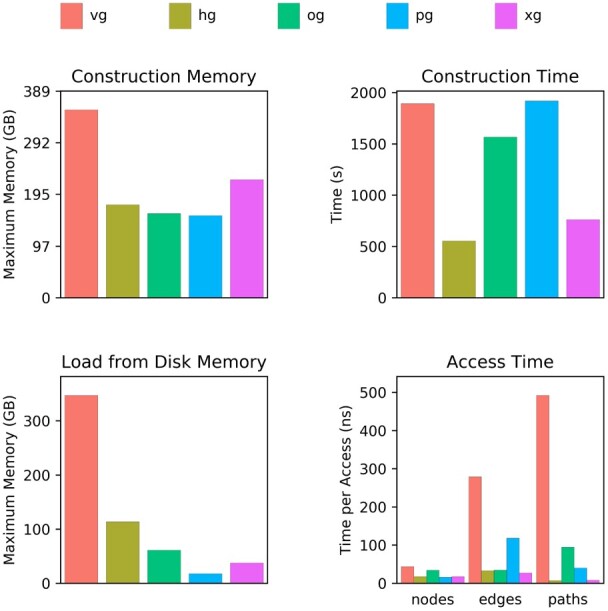
Performance on a graph of structural variants from the HGSVC. Abbreviations used here and in subsequent figures and tables: vg, VG; hg, HashGraph; og, ODGI; pg, PackedGraph; xg, XG. All four new graph implementations compare favorably to VG. PackedGraph tends to be the most memory efficient, HashGraph tends to be the fastest, and ODGI is balanced in between. XG provides good performance on both memory usage and speed, but it is static
