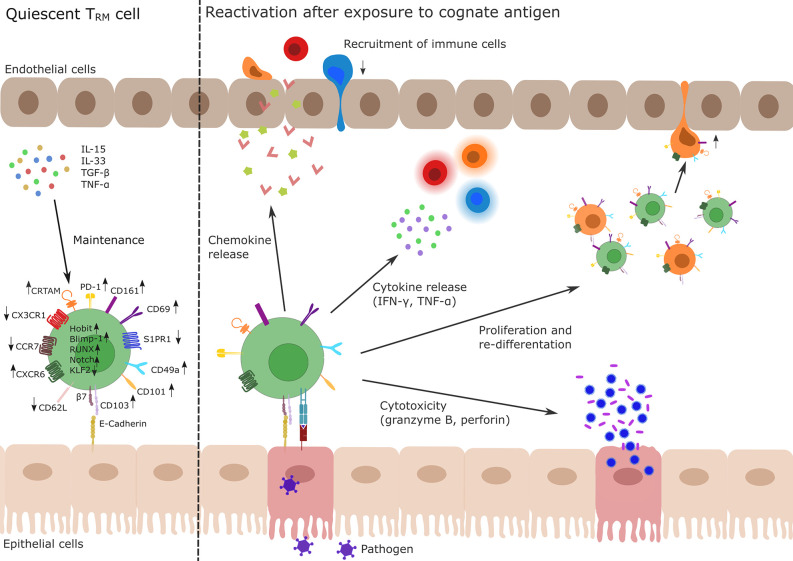
Figure 1.
Profile and function of TRM cells. Left side: TRM cells develop during primary infection. The differentiation and maintenance of TRM cells is controlled by tissue-derived signals, e.g., TNF-α, TGF-β or IL-15 and IL-33 resulting in the up- and down-regulation of different genes via activity of the transcription factors Hobit, Blimp-1, Runx3, and Notch and the silencing of Klf2. In particular, upregulation of CD69 and CD103 and simultaneous downregulation of S1PR1 are key drivers of TRM cell tissue retention. Other membrane molecules highly expressed in TRM cells are CD49a, CD101, PD-1, CRTAM, and CXCR6 while CD62L, CCR7, and CX3CR1 show a decreased expression pattern in TRM cells. Right side: After re-exposure to a cognate antigen (e.g., from a pathogen, shown in purple), TRM cells are able to initiate a fast immune response. This includes chemokine release to recruit lymphocytes (indicated as red, orange, and blue immune cells) to the site of infection, release of pro-inflammatory cytokines (IFN-γ, TNF-α) to activate other cells as well as the production of the cytotoxic effectors perforin or granzyme B. There is also evidence for the ability of TRM cells to proliferate or to re-differentiate (indicated as green and orange cells) and to leave the tissue (orange ex-TRM cells; for details cf. main text). TRM, tissue-resident memory T cell; TNF, tumor necrosis factor; TGF, transforming growth factor; IL, Interleukin; KLF, Krüppel-like factor; CD, cluster of differentiation; S1PR1, sphingosine-1-phosphate receptor 1; PD-1, programmed cell death protein 1; CRTAM, cytotoxic and regulatory T-cell molecule; CXCR, CXC-motif chemokine receptor; CCR, Chemokine receptor.