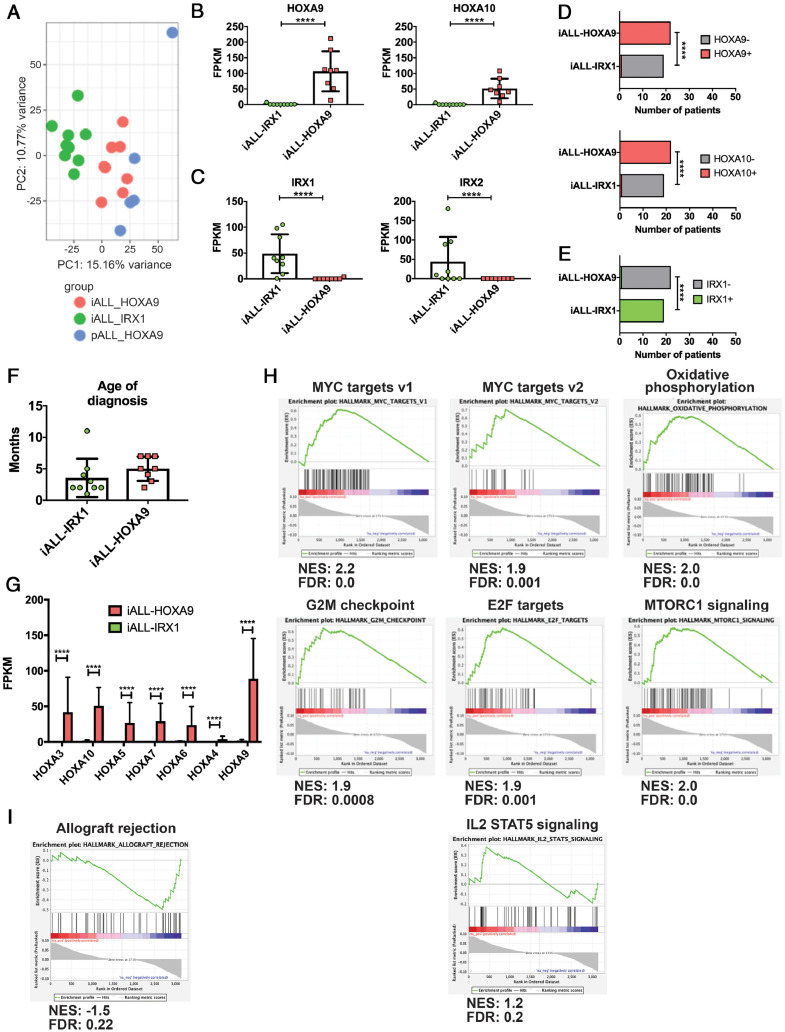
Figure 1.
HOXA9/HOXA10–IRX1 expression defines two subgroups of infant MLL-AF4-driven ALL (Andersson et al.[9] data set). (A) PCA of patients defined by HOXA9/HOXA10 and IRX1 expression and age at diagnosis. Green = infants with IRX1 expression, pink = infants with HOXA9/HOXA10 expression, blue = pediatric patients. All pediatric patients expressed HOXA9. (B)HOXA9 and HOXA10 expression in the two subgroups. RNA Sequencing data are expressed as means ± SD; each dot represents a sample. (C)IRX1 and IRX2 expression in the two subgroups. RNA sequencing data are expressed as means ± SD; each dot represents a sample. (D) Fisher's exact test comparing patient samples based on HOXA9 and HOXA10 expression levels (samples were deemed negative at FPKM <1 and positive at FPKM >1). (E) Fisher's exact test comparing patient samples based on IRX1 expression levels (samples were deemed negative if FPKM was <1 and positive if FPKM was >1). (F) Age at diagnosis of patients with iALL-IRX1 and iALL-HOXA9. Data are expressed as means ± SD. Student's t test was performed. (G) Expression of HOXA cluster genes in the two subgroups. Data are expressed as means + SD. (H,I) Gene set enrichment analysis of iALL-IRX1 and iALL-HOXA9 patient samples. iALL-IRX1 patients (H) exhibit enrichment for MYC targets, oxidative phosphorylation, G2M checkpoints, E2F targets, MTORC1, and IL2 STAT5 signaling, whereas iALL-HOXA9 (I) patient samples exhibit enrichment for allograft rejection. FPKM=fragments per kilobase of transcript per million; FDR=false discovery rate; NES=normalized enrichment score. ****p < 0.0001.