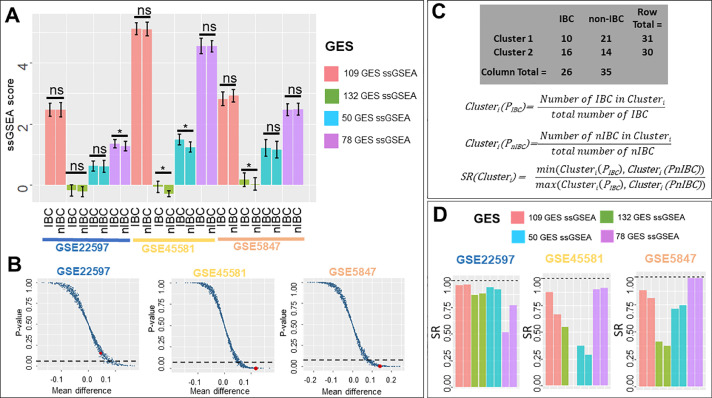
Fig. 3.
ssGSEA scores of IBC gene signatures. A) ssGSEA scores in IBC and nIBC samples across three datasets (* p< 0.05, two-tailed Student's t-test; error bars represent standard deviation). B) Scatter plot showing ssGSEA scores calculated from 1000 random combinations of 132 genes each. Each data point in the plot represents data for a randomly generated gene list; the single point highlighted in red is calculated from 132 GES. x-axis is Mean difference = mean(ssGSEAIBC) - mean(ssGSEAnIBC) and y-axis is p-value estimated by two-tailed Student's t-test of ssGSEA score across IBC and nIBC group (dotted lines denote p-value = 0.05). C) SR (sample ratio) method to measure accuracy of clustering based on ssGSEA scores (GSE5847 clusters) D) SR (sample ratio) of different gene signature ssGSEA scores across three datasets.