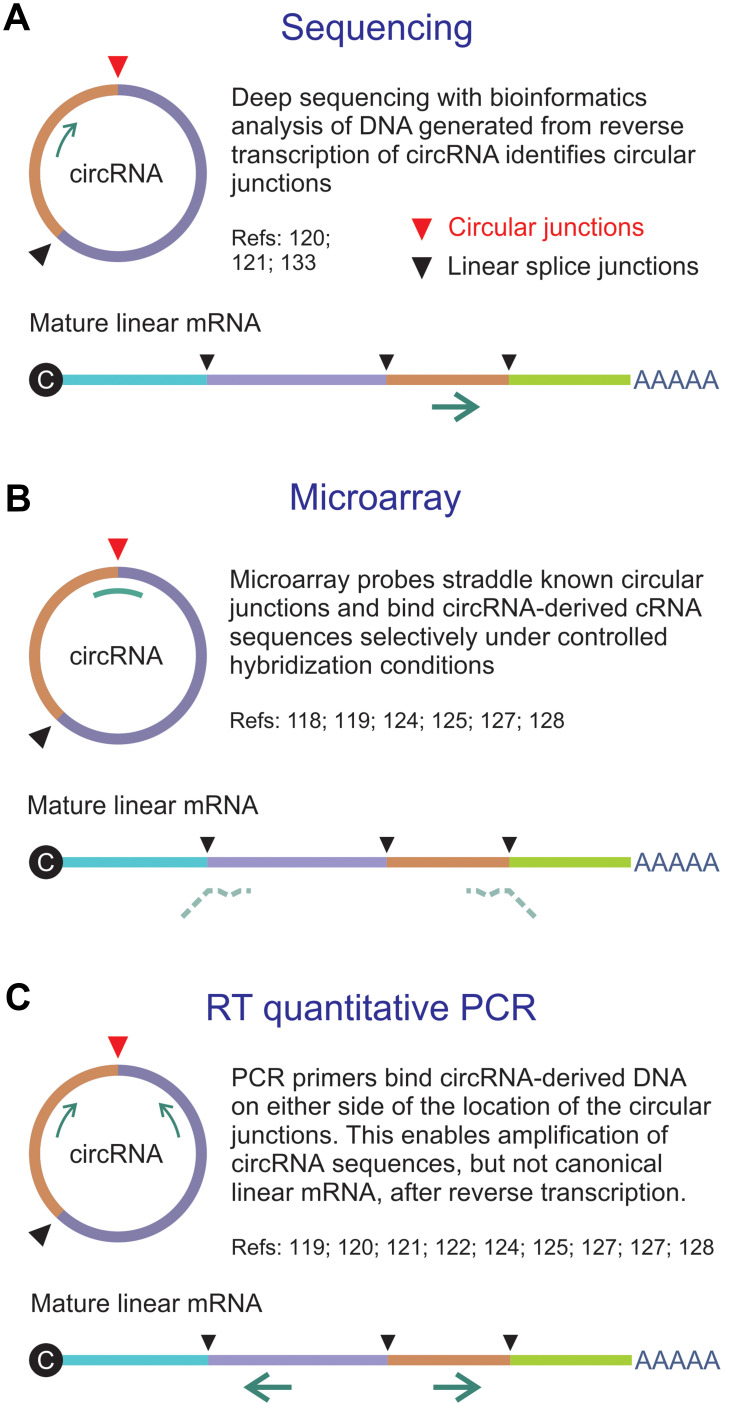
Figure 2.
Methods for assay of circRNA. (A) Direct sequencing of a reverse transcribed circRNA typically employs a primer that is complementary to an exon. The sequencing reaction does not discriminate between linear RNA and circRNA, but bioinformatics analysis allows identification of circular junctions (red arrowhead). (B) Microarrays make use of probes that straddle unique and defined circularization junctions. Typically random primers are used to generate labeled cRNAs from circRNA templates. Probes span the circular junctions and hybridize to the cRNAs with higher Tms than the partly complementary sequences of linear mature RNA. (C) Reverse transcriptase (RT) quantitative PCR entails specific amplification of sequences derived from circRNAs by using primers that flank the circular junctions. The configuration of the amplifying primers is such that mature linear mRNA is not amplified.