Fig. 1.
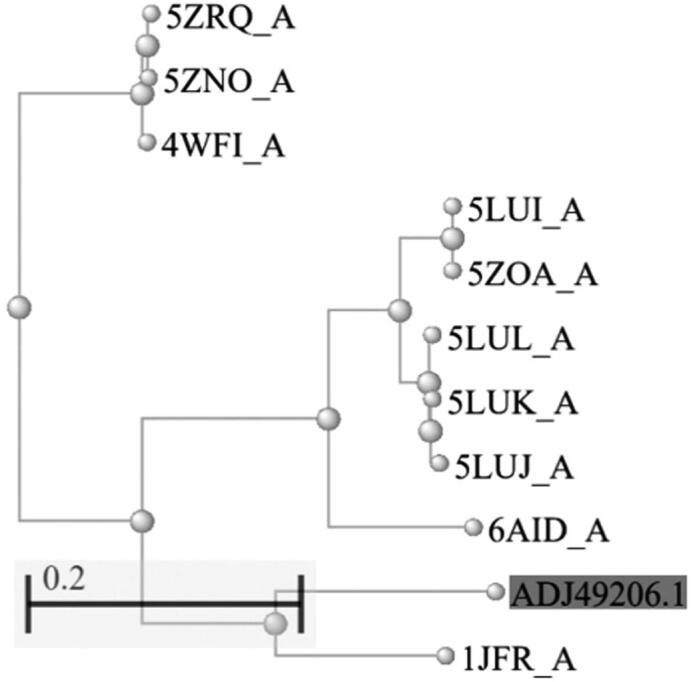
Phylogenetic tree of AML (highlighted in grey; GenBank ID: ADJ49206.1), Saccharomonospora viridis cutinase S176A/S226P/R228S mutant (PDB ID: 5ZRQ_A), Saccharomonospora viridis cutinase S176A/S226P/R228S mutant (PDB ID: 5ZNO_A), with Saccharomonospora viridis cutinase (PDB ID: 4WFI_A), Thermobifida cellulosilytica cutinase 1 (PDB ID: 5LUI_A), BTA_hydrolase 1 from Thermobifida fusca (PDB ID: 5ZOA_A), Thermobifida cellulosilytica cutinase, triple variant (PDB ID: 5LUL_A), Thermobifida cellulosilytica cutinase 2, double variant (PDB ID: 5LUK_A), Thermobifida cellulosilytica cutinase 2 (PDB ID: 5LUJ_A), Thermobifida alba cutinase Est119 (PDB ID: 6AID_A), and Streptomyces exfoliatus lipase (PDB ID: 1JFR_A). The tree of the first 10 BLASTp hits was constructed using pairwise alignment in BLASTp [16] with the setting of “Fast Minimum Evolution” tree method, “Grishin (protein)” distance model, and a maximum sequence distance of 0.4.
