Abstract
Hypopharyngeal squamous cell carcinoma (HSCC) remains one of the most lethal malignancies in the head and neck. Long noncoding RNA (lncRNA) HOXA11-AS is proven to function as an oncogene and a therapeutic target in various tumors. Our previous study and others have demonstrated that HOXA11-AS is one of the most upregulated lncRNAs in HSCC. However, the role of HOXA11-AS in HSCC has not yet been identified. The current study demonstrated that the expression of HOXA11-AS was significantly upregulated in HSCC tumors and was positively associated with lymph node metastasis. Moreover, functional experiments revealed that HOXA11-AS knockdown suppressed the proliferation and migration potential in FaDu cells. Furthermore, luciferase reporter gene assay combined with cellular functional experiments demonstrated that HOXA11-AS functioned as a molecular sponge for miR-155, and inhibition of miR-155 attenuated the suppressive effect of HOXA11-AS knockdown on the aggressive phenotype in HSCC. This study identifies a tumor-promoting role of HOXA11-AS in HSCC and suggests HOXA11-AS might be a potential diagnostic and therapeutic target for HSCC.
Key words: Hypopharyngeal squamous cell carcinoma (HSCC), Long noncoding RNAs (lncRNAs), HOXA11-AS, miR-155, Proliferation, Migration
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) refers to a large heterogeneous group of tumors arising from the epithelium of the oral cavity, oropharynx, larynx, or hypopharynx1. Hypopharyngeal squamous cell carcinoma (HSCC) is the most dismal HNSCC, with a poor 5-year survival rate ranging from 30% to 50%2. HSCC is characterized by a high incidence of metastasis to the draining lymph nodes3,4, which has been considered the most potent prognostic factor and responsible for a decrease in the overall survival rate by at least 25%5. In addition, cervical lymph node metastasis is recognized as an important predictive indicator of early regional recurrence and distant metastasis3,5,6, which also negatively impact the prognosis for HSCC patients3,7. Despite advances in functional outcomes, the overall survival for HSCC patients remains unimproved over the last decades8. There is an unmet need to study the molecular mechanisms of HSCC initiation and progression and to identify novel molecular biomarkers that can predict lymph node metastasis and serve as therapeutic targets for patients with HSCC.
Long noncoding RNAs (lncRNAs) play essential roles in tumor initiation and progression through activating oncogenes or blocking tumor suppressors via different mechanisms, including miRNA decoy, mRNA alternative splicing, epigenetic and posttranslational regulation, and chromatin modification9–11. Multiple studies demonstrated that cytoplasmic lncRNAs may serve as sponges that sequester microRNAs to manipulate other RNAs’ expression, which is the competing endogenous RNA (ceRNA) interactions9,12,13. Moreover, several studies have revealed that lncRNA expression is dysregulated in multiple types of cancers, and some lncRNAs are associated with cancer recurrence and poor prognosis14,15. Our previous work, aiming to study the lncRNAs in HSCC utilizing the Arraystar Human LncRNA Microarray to screen for differentially expressed lncRNAs in three paired HSCC tumor specimens and matched normal controls, identified two novel lncRNAs, AB209630 and AB01956216. In addition, multiple lncRNAs are also dysregulated in HSCC but fail to reach the rigorous criteria, which may be attributed to the small sample size and the heterogeneity of HSCC16. lncRNA HOXA11 antisense RNA (HOXA11-AS) turns out to be one of the top candidate genes that possess HSCC-specific expression pattern and relatively high expression levels in HSCC tumors.
HOXA11-AS has recently emerged as an important tumor-promoting lncRNA in multiple cancers including gastric cancer17, breast cancer18, and non-small cell lung cancer19. HOXA11-AS can promote cell growth and invasion of gastric cancer through scaffolding PRC2, LSD1, and DNMT1, or sponging miR-129717. Of note, the study by Yao and colleagues indicated that HOXA11-AS may be a potential circulating biomarker for early detection of squamous cell carcinomas that originate in the oral cavity, oropharynx, larynx, and hypopharynx20; however, the functional roles and the underlying mechanism of HOXA11-AS were not elucidated in their work.
Therefore, our study aims to validate the expression pattern of HOXA11-AS in HSCC tumor versus adjacent normal tissues, to explore its correlation with clinical features of HSCC, and to identify the functional roles and the underlying mechanism of HOXA11-AS in HSCC FaDu cells in vitro. Our findings suggest that HOXA11-AS serves as a tumor-promoting lncRNA and a potential therapeutic target of HSCC.
MATERIALS AND METHODS
Patients and Specimens
Eighteen paired HSCC tumor specimens and adjacent normal tissues were collected from HSCC patients who underwent surgical resection from June 2014 to June 2015 in Qilu Hospital of Shandong University. None of these patients had received neoadjuvant chemotherapy or radiotherapy before surgeries. All these collected specimens were immediately frozen and stored in liquid nitrogen until use. Retrospective clinicopathological characteristics of the patients were also obtained and listed in Table 1, including gender, age, tumor size, differentiation, T stage, lymph node metastasis, and clinical stage. Written informed consent was obtained from each individual before tissue collection. The protocol was approved by the Institutional Research Ethics Committee of Qilu Hospital of Shandong University.
Table 1.
Clinicopathological Characteristics of the Patients
| HOXA11-AS1 Expression | p Value | ||
|---|---|---|---|
| Low | High | ||
| Gender | 0.303 | ||
| Male | 8 | 9 | |
| Female | 1 | 0 | |
| Age | 55.67 ± 1.691 | 58.44 ± 0.7658 | 0.154 |
| Size (cm) | 3.067 ± 0.4481 | 4.378 ± 0.5456 | 0.082 |
| Nuclear grade | 0.324 | ||
| G1 | 2 | 0 | |
| G2 | 3 | 4 | |
| G3 | 4 | 5 | |
| T stage | 0.392 | ||
| T1 | 3 | 1 | |
| T2 | 1 | 1 | |
| T3 | 5 | 5 | |
| T4 | 0 | 2 | |
| Lymph node meta | 0.014 | ||
| N0 | 6 | 0 | |
| N1 | 2 | 2 | |
| N2 | 0 | 1 | |
| N3 | 1 | 6 | |
| Distant meta | NA | ||
| M0 | 9 | 9 | |
| Clinical stage | 0.014 | ||
| I | 2 | 0 | |
| III | 6 | 2 | |
| IV | 1 | 7 | |
Cell Culture and Reagents
The human HSCC cell line FaDu was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). FaDu cells were derived from primary squamous cell carcinoma of the hypopharynx and cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Hyclone Laboratories, Logan, UT, USA) with 10% (v/v) fetal bovine serum (FBS; Gibco, Gaithersburg, MD, USA) at 37°C in a humidified atmosphere containing 5% (v/v) CO2.
Small Interfering RNA Transfection
FaDu cells were transfected with the small interfering RNAs (siRNAs) targeting HOXA11-AS or negative control. The sequence of HOXA11-AS siRNA was 5′-AGG CCAAGUCCGAGUUCCAUUUCUU-3′, and the sequence of control siRNA was 5′-UUCUCCGAACGUGUCACGUUU-3′ (Ribobio, Guangzhou, China). FaDu cells were treated with these siRNAs at the concentration of 50 nM, utilizing Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
RNA Isolation, Reverse Transcription Polymerase Chain Reaction (PCR), and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from HSCC specimens and FaDu cells utilizing TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the protocol, and 2 μg of total RNA was reversely transcribed into cDNA with the aid of M-MLV reverse transcriptase (TaKaRa Biotechnology, Dalian, China). Subsequently, qRT-PCR assays were conducted using SYBR Green kit (Applied Biosystems, Foster City, CA, USA) in the Mastercycler ep realplex real-time PCR system (Eppendorf, Hamburg, Germany). The primer sequences were as follows: HOXA11-AS, 5′-AGCAACAGATCGTCACTCGG-3′ (forward) and 5′-GAGAACGAGGACCCTGCAAT-3′21 (reverse); GADPH, 5′-AGAAGGCTGGGGCTCATTTG-3′ (forward) and 5′-AGGGGCCATCCACAGTCTTC-3′ (reverse). The relative expression levels were calculated using the 2−ΔΔCt method.
Subcellular Fractionation
The subcellular fraction of nuclear/cytoplasmic expression of HOXA11-AS in FaDu cells was examined using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. The relative amount of nuclear/cytoplasmic expression of HOXA11-AS was detected by qRT-PCR.
Dual-Luciferase Reporter Assay
The plasmids with the HOXA11-AS sequence containing the predicted miR-155 binding site or the mutant miR-155 binding site were generated using the dual-luciferase miRNA Target Expression vector pmirGlo kit (Promega, Madison WI, USA). HEK293 cells were seeded in six-well plates overnight and were cotransfected with the pmirGLO-HOXA11-AS-WT or -MUT reporter plasmids (both 500 ng) and the mimic miR-155 or the negative control (both 30 nM). Relative luciferase activity normalized to the Renilla luciferase activity was measured 24 h after the cotransfection using the dual-luciferase reporter assay system according to the manufacturer’s instructions.
Cell Proliferation Assay
A total of 5.0 × 103 FaDu cells were seeded into each well of 96-well plates and cultured for 0, 24, 36, 48, and 60 h after transfection with HOXA11-AS or control siRNA. At each indicated time point, cells were counted using the cell Countstar (IC1000). The curve of cell proliferation was drawn. Experiments were performed in triplicate.
Wound Healing Assay
FaDu cells (5.0 × 104) were plated into each well of six-well plates and transfected with HOXA11-AS or control siRNAs. After 48 h, the confluent monolayer was scraped with a sterile 200-μl pipette tip to create cell-free areas. After two washes with phosphate-buffered saline, medium without FBS was added and cells were incubated for 24 h. Images were captured at 0 and 24 h after scratching and analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The percentage of wound closure was quantified by dividing the healing wound width at 24 h by the initial one at 0 h as previously described.22
FluoroBlok Transwell Migration Assays
The FluoroBlok Transwell migration assays were conducted using the FluroBlok Cell Culture Inserts (Corning, Corning, NY, USA) according to the manufacturer’s instruction. Briefly, after rehydration of the FluroBlok membrane, cell suspensions (1.2 × 105 cells/200 μl, FBS-free medium) were transferred to the upper chambers, and 600 μl of medium with 10% FBS was added to the bottom chambers. After incubation at 37°C, 5% CO2 for 60 h, the inserts were transferred to a 24-well plate containing 500 μl of 4 μg/ml Calcein AM (Invitrogen) in Hanks buffered saline, and incubated for another 1 h. The pictures of the inserts were then taken by an inverted fluorescence microscope (Olympus, Tokyo, Japan). The penetrated cells were counted in four randomly selected high-microscopic fields (400×).
Gene Set Enrichment Analysis
The RNA sequencing data of HSCC were acquired from The Cancer Genome Atlas Research Network (TCGA; http://cancergenome.nih.gov ) to generate the ranked list of genes for which HOXA11-AS is positively associated. The Preranked Gene Set Enrichment Analysis23 was run using the curated gene sets from the Molecular Signatures Database (MSigDB), Broad Institute.
Statistical Analysis
Statistical analyses were carried out utilizing the SPSS version 17.0 (IBM, USA) and GraphPad Prism version 5.0 (GraphPad software, USA). The differential expression of HOXA11-AS in HSCC specimens was assessed using the paired t-test. The correlation of HOXA11-AS expression with clinicopathological parameters was analyzed using the chi-squared or Pearson correlation test. Differences between the experimental groups and control groups were assessed by Student’s t-test. Data were presented as the mean ± standard error of the mean (SEM). A value of p < 0.05 was considered as statistically significant.
RESULTS
HOXA11-AS Expression Was Upregulated and Correlated With Lymph Node Metastasis in HSCC
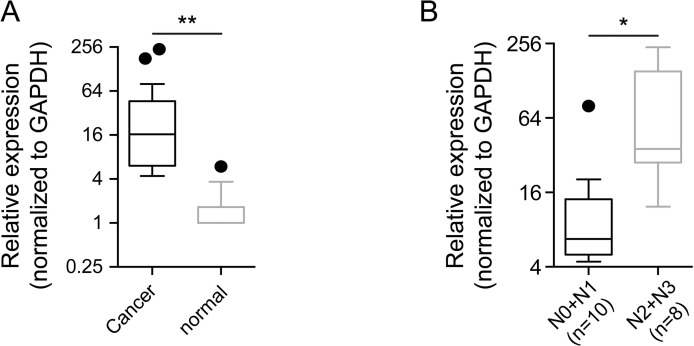
Our previous screening effort for exploration of differentially expressed lncRNAs in HSCC utilizing the Arraystar Human LncRNA Microarray identified HOXA11-AS as one of the top candidate lncRNAs. The expression level of HOXA11-AS in 18 pairs of HSCC tumor specimens and adjacent normal tissues was detected and analyzed using qRT-PCR. Our results suggested that HOXA11-AS expression was remarkably upregulated in human HSCC tumors compared with the adjacent normal tissues (p = 0.0082) (Fig. 1A). The clinicopathological characteristics of the 18 HSCC patients are shown in Table 1. High expression of HOXA11-AS was found to be significantly associated with lymph node metastasis (p = 0.014) and advanced clinical stage (p = 0.014). The expression of HOXA11-AS in the advanced lymph node metastatic group (N2 + 3) was significantly elevated compared with the less advanced lymph node metastatic group (N0 + 1) (p = 0.0334) (Fig. 1B). These findings suggest a potential role of HOXA11-AS as a tumor-promoting lncRNA in HSCC progression.
Figure 1.
HOXA11-AS expression was upregulated in hypopharyngeal squamous cell carcinoma (HSCC). (A) The expression of HOXA11-AS was significantly upregulated in HSCC tumor specimens compared with the adjacent normal tissues (p = 0.0082, n = 18 pairs). (B) The expression of HOXA11-AS in the advanced lymph node metastatic group (N2 + 3, n = 8) was significantly higher than in the less advanced lymph node metastatic group (N0 + 1, n = 10; p = 0.0334). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. Comparison was analyzed by paired t-test and unpaired t-test, respectively. *p < 0.05; **p < 0.01.
Knockdown of HOXA11-AS Inhibited the Aggressive Phenotype of HSCC FaDu Cells
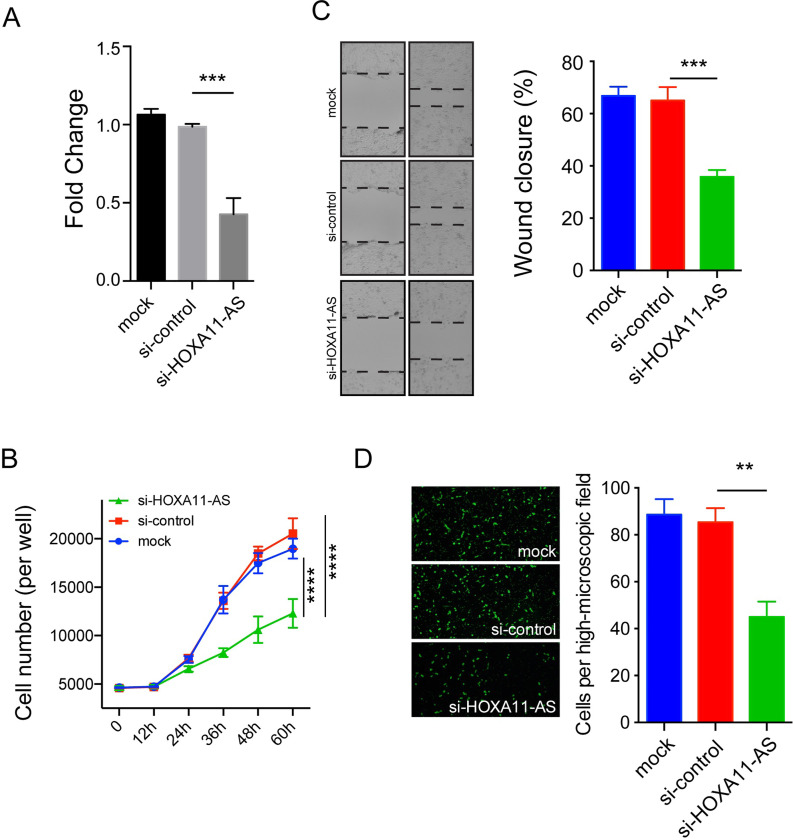
To primarily explore the tumor-promoting effect of HOXA11-AS in HSCC progression, we utilized a loss-of-function strategy. The HOXA11-AS expression was downregulated more than 60% by HOXA11-AS siRNA in comparison with the control siRNA or the mock group (Fig. 2A). Cell proliferation assay revealed that knockdown of HOXA11-AS remarkably inhibited cell proliferation in FaDu cells (p < 0.0001) (Fig. 2B). Wound healing assay showed that significantly less wound closure was observed in the HOXA11-AS siRNA group compared with the control siRNA or mock groups (p = 0.0009) (Fig. 2C). Fluoroblok Transwell migration assay demonstrated that the number of cells that penetrated the membrane was significantly reduced in the HOXA11-AS siRNA group compared with the control siRNA or mock groups (p = 0.0014) (Fig. 2D). Taken together, in vitro functional assays indicate a tumor-promoting effect of HOXA11-AS in HSCC.
Figure 2.
HOXA11-AS knockdown suppressed cell proliferation and migration of FaDu cells. (A) The HOXA11-AS expression was significantly downregulated by HOXA11-AS-specific siRNA compared with the control siRNA group (p = 0.0015). (B) HOXA11-AS knockdown significantly suppressed cell proliferation in FaDu cells (p < 0.0001). HOXA11-AS knockdown significantly inhibited cell migration in FaDu cells as determined by wound healing assay (p = 0.0009) (C) and Fluoroblok Transwell migration assay (p = 0.0014) (D). Data are presented as the mean ± standard error of the mean (SEM). All the assays were performed in triplicate. **p < 0.01, ***p < 0.001, ****p < 0.0001.
HOXA11-AS Knockdown Inhibited Cell Proliferation and Migration by Sponging miR-155
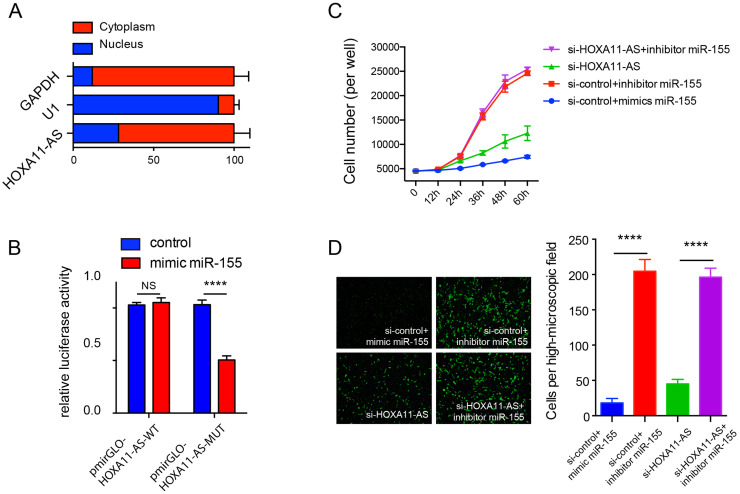
The distribution of HOXA11-AS in FaDu cells was examined using subcellular fractionation followed by qRT-PCR, and the result revealed that HOXA11-AS was mainly localized in cytoplasm (Fig. 3A). The Gene Set Enrichment Analysis using the cell component modules showed that HOXA11-AS was functionally associated with the ribosome subunit (see Supplemental Fig. 1A, available at https://pan.baidu.com/s/1N3BlDvPbMQAokP-MO3KefQ ). Considering the cytoplasmic subcellular localization of HOXA11-AS, it is possible that the lncRNA exerted a tumor-promoting effect by acting as a molecular sponge to block specific target miRNAs. Interestingly, the tumor suppressor miR-155 was found to be a potential target of HOXA11-AS by DIANA-LncBase v2, as well as the GSEA (see Supplemental Fig. 1B, available at https://pan.baidu.com/s/1N3BlDvPbMQAokP-MO3KefQ ). To verify the binding between HOXA11-AS and miR-155, we performed dual-luciferase reporter assay using the HOXA11-AS sequence harboring the putative miR-155 binding sites (pmirGLO-HOXA11-AS-WT), as well as a mutated HOXA11-AS sequence (pmirGLO-HOXA11-AS-MUT) (see Supplemental Fig. 1C, available at https://pan.baidu.com/s/1N3BlDvPbMQAokP-MO3KefQ ). The results showed that the relative luciferase activity of the mimic miR-155 group was significantly reduced compared with the control group in the pmirGLO-HOXA11-AS-WT group (p = 0.0009) (Fig. 3B), but not in the pmirGLO-HOXA11-AS-MUT group. Moreover, cell proliferation assay revealed that mimic miR-155 significantly suppressed cell proliferation, while inhibitor miR-155 significantly promoted cell proliferation in FaDu cells (p < 0.0001) (Fig. 3C). The suppressive effect on cell proliferation of HOXA11-AS knockdown was reversed by adding inhibitor miR-155 (p < 0.0001) (Fig. 3C). Fluoroblok Transwell migration assay demonstrated that the penetrated cell numbers were significantly increased in the inhibitor miR-155 group compared with the mimic miR-155 group (p < 0.0001) (Fig. 3D), and the suppressive effect on cell migration of HOXA11-AS knockdown was attenuated by adding inhibitor miR-155 (p < 0.0001) (Fig. 3D). These findings suggest that the HOXA11-AS exerts the tumor-promoting effect via sponging miR-155 in HSCC.
Figure 3.
HOXA11-AS exerted the tumor-promoting role via sponging the tumor suppressor miR-155. (A) Subcellular fractionation followed by quantitative real-time polymerase chain reaction (qRT-PCR) revealed that HOXA11-AS was mainly localized in the cytoplasm of FaDu cells. (B) The relative luciferase activity of the HEK293 cells cotransfected with pmirGLO-HOXA11-AS-WT or pmirGLO-HOXA11-AS-MUT and mimic miR-155 or negative control. (C) Cell proliferation assay revealed that mimic miR-155 significantly suppressed cell proliferation, while inhibitor miR-155 significantly enhanced cell proliferation, and the suppressive effect of HOXA11-AS knockdown on cell proliferation was reversed by adding inhibitor miR-155 in FaDu cells. (D) Fluoroblok Transwell migration assay revealed that mimic miR-155 significantly suppressed cell migration, while inhibitor miR-155 significantly enhanced cell migration, and the suppressive effect of HOXA11-AS knockdown on cell migration was reversed by adding inhibitor miR-155 in FaDu cells (all p < 0.0001). Data are presented as the mean ± SEM. All the assays were performed in triplicate. ****p < 0.0001.
DISCUSSION
lncRNAs emerges as key regulators and potential therapeutic targets in cancers including HNSCC24. HOTAIR, one of the first characterized lncRNAs, is demonstrated to be upregulated in HNSCC and to promote cell migration and invasion through miR-7-dependent HuR regulation25. MALAT1 upregulation is frequently observed in several types of head and neck cancers26–28 and promotes tongue cancer cell growth and metastasis by regulating JAG1 expression via acting as a ceRNA for miR-12426. However, the role of lncRNAs in HSCC pathophysiology still remains largely unknown. Our previous study aimed to explore the biological function of lncRNAs, as well as their prognostic value in HSCC clinical management. As a preliminary screening effort, our group performed global profiling of human lncRNAs in three paired HSCC tumor specimens and the matched normal controls using the Arraystar Human LncRNA Microarray, and identified two novel lncRNAs, AB019562 and AB209630, which were subsequently proven to play critical roles in HSCC cell proliferation and invasion16. Moreover, multiple genes are also found be to dysregulated in HSCC but fail to reach statistical significance, partially due to the small sample size and the heterogeneity of HSCC. HOXA11-AS turns out to be one of the top candidate genes that are upregulated in HSCC tumor specimens relative to matched normal controls, and at the same time exhibits a high expression level in tumor specimens. Thus, the present study aimed to identify the expression pattern and the functional roles of HOXA11-AS in HSCC.
The expression of HOXA11-AS was found to be significantly upregulated in HSCC tumor specimens compared with the adjacent normal tissues, and the high expression of HOXA11-AS was significantly associated with lymph node metastasis of HSCC. Several studies demonstrated that HOXA11-AS is dysregulated in a variety of cancer types17–19,29–31, including gastric cancer17 and serous ovarian cancer29 among others, and its expression is associated with the poor clinicopathological parameters17,29,30,32. The above findings indicate that HOXA11-AS might function as an oncogene in a variety of cancers, including HSCC.
Notably, HOXA11-AS expression is quite high in HSCC tumor specimens, particularly in cases with the advanced lymph node metastasis, which might gain its value as a diagnostic biomarker that could be detected by the minimally invasive manners such as blood sampling. In support of our findings, Yao and colleagues20 found HOXA11-AS could serve as a potential circulating biomarker in HNSCC, due to its upregulation and high stability in plasma, which was further validated in a large cohort. One limitation of our present study is that we did not check the HOXA11-AS in patients’ plasma due to the lack of blood samples of HSCC patients with follow-up information, which warrants further investigation.
Multiple studies demonstrate that HOXA11-AS plays critical roles in regulating the aggressive phenotypes of various tumor cells17,19,21,31,33–35. Liu et al. demonstrated that HOXA11-AS knockdown impairs the aggressive capacities in vitro and in vivo and induces the G0/G1 arrest of cell cycle via regulating β-catenin and KLF2 in gastric cancer cells21. Moreover, HOXA11-AS promotes cell proliferation and invasion and triggers the epithelial–mesenchymal transition in hepatocellular carcinoma cells31. Our data also revealed that knockdown of HOXA11-AS suppressed the proliferation and migration potential in HSCC FaDu cells, which supported the abovementioned findings that high expression of HOXA11-AS was associated with lymph node metastasis in HSCC patients. Together, our results suggest that HOXA11-AS may serve as an oncogene in HSCC, which warrants further study to explore its tumor-promoting effect in vivo.
To study the underlying mechanism of the oncogene HOXA11-AS, we first investigated its subcellular localization in FaDu cells and demonstrated that HOXA11-AS mainly localized in the cytoplasm. Recently, a number of studies have demonstrated that the cytoplasmic lncRNAs can regulate the malignant progress by serving as a sponge to sequester its target miRNAs, which is the ceRNA mechanism9,12,13. Sun et al. showed that HOXA11-AS acts as a ceRNA for miR-1297 to impose posttranscriptional regulation in gastric cancer cells17. Zhan et al. demonstrated that HOXA11-AS sponges miR-214-3p to facilitate hepatocellular carcinoma progression31. Based on the cytoplasmic localization of HOXA11-AS, we hypothesize that HOXA11-AS may also act as a molecular sponge for specific miRNA in HSCC. To investigate this hypothesis, we analyzed the database from DIANA-LncBase v2 and the GSEA and found that miR-155 was a potential target for HOXA11-AS. Furthermore, dual-luciferase reporter assays showed that miR-155 could directly bind to HOXA11-AS. In addition, miR-155 inhibition attenuated the suppressive effect on cell proliferation and migration in FaDu cells transfected with HOXA11-AS siRNA. Dysregulated miR-155 plays an important role in various tumors36–42, including HNSCC43–46; however, the biological function of miR-155 in HNSCC tumorigenesis is controversial43,45–47, which may be attributed to multiple factors such as ethnic diversity and tumor heterogeneity. Downregulation of miR-155 is found to be associated with the lymph node metastatic disease in oral squamous cell carcinoma; in contrast, upregulation of miR-155 promotes proliferation and invasion in laryngeal squamous cell carcinoma through targeting SOCS1 and STAT348. In our study, we demonstrated that inhibitor miR-155 enhanced FaDu cell proliferation and migration in vitro, whereas mimic miR-155 inhibited the aggressive phenotype, suggesting that miR-155 functioned as a tumor suppressor in HSCC. Lerner and colleagues reported that miR-155 dramatically inhibits cell proliferation and migration potential in HNSCC FaDu and UM-SCC-1 cell lines46. Additionally, they also demonstrated that miR-155 is downregulated in HNSCC tumor tissues, and its downregulation in blood samples of HNSCC patients is correlated with distant metastasis46, which supports our finding that miR-155 serves as a tumor suppressor in HSCC. Collectively, our data suggest that HOXA11-AS functions as a tumor-promoting lncRNA through sponging the tumor suppressor miR-155 in HSCC. Our study further expanded the ceRNA profile of HOXA11-AS in the context of malignancies; however, one limitation of the current study is that we did not check the biological function of the proven HOXA11-AS-target miRNAs in HSCC, which warrants further studies.
CONCLUSION
In summary, our study demonstrates that the lncRNA HOXA11-AS is upregulated in HSCC tumors, and the upregulation is significantly correlated with the advanced lymph node metastasis. Moreover, HOXA11-AS functions as a tumor-promoting lncRNA in HSCC progression via sponging miR-155. Overall, our work suggests that HOXA11-AS may serve as a therapeutic target for HSCC and encourages further investigation for its diagnostic value, particularly as a circulating biomarker in HSCC.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Nos. 81802577 and 81702521) and Provincial Natural Science Foundation of Shandong (Nos. ZR2018BH018 and ZR2017PH019).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gourin CG, Johnson JT. A contemporary review of indications for primary surgical care of patients with squamous cell carcinoma of the head and neck. Laryngoscope 2009;119:2124–34. [DOI] [PubMed] [Google Scholar]
- 3. Spector JG, Sessions DG, Haughey BH, Chao KS, Simpson J, El Mofty S, Perez CA. Delayed regional metastases, distant metastases, and second primary malignancies in squamous cell carcinomas of the larynx and hypopharynx. Laryngoscope 2001;111:1079–87. [DOI] [PubMed] [Google Scholar]
- 4. Joo YH, Cho KJ, Kim SY, Kim MS. Prognostic significance of lymph node density in patients with hypopharyngeal squamous cell carcinoma. Ann Surg Oncol. 2015;22(Suppl 3):S1014–9. [DOI] [PubMed] [Google Scholar]
- 5. Hall SF, Groome PA, Irish J, O’Sullivan B. The natural history of patients with squamous cell carcinoma of the hypopharynx. Laryngoscope 2008;118:1362–71. [DOI] [PubMed] [Google Scholar]
- 6. Werner JA, Dunne AA, Myers JN. Functional anatomy of the lymphatic drainage system of the upper aerodigestive tract and its role in metastasis of squamous cell carcinoma. Head Neck 2003;25:322–32. [DOI] [PubMed] [Google Scholar]
- 7. Omura G, Ando M, Saito Y, Kobayashi K, Yamasoba T, Asakage T. Disease control and clinicopathological prognostic factors of total pharyngolaryngectomy for hypopharyngeal cancer: A single-center study. Int J Clin Oncol. 2015;20:290–7. [DOI] [PubMed] [Google Scholar]
- 8. Kim JW, Kim MS, Kim SH, Kim JH, Lee CG, Kim GE, Keum KC. Definitive chemoradiotherapy versus surgery followed by adjuvant radiotherapy in resectable stage III/IV hypopharyngeal cancer. Cancer Res Treat. 2016;48:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmitt AM, Chang HY. Long Noncoding RNAs: At the intersection of cancer and chromatin biology. Cold Spring Harb Perspect Med. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bolha L, Ravnik-Glavac M, Glavac D. Long noncoding RNAs as biomarkers in cancer. Dis Markers 2017;2017:7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer 2016;139:269–80. [DOI] [PubMed] [Google Scholar]
- 14. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shukla S, Zhang X, Niknafs YS, Xiao L, Mehra R, Cieslik M, Ross A, Schaeffer E, Malik B, Guo S, Freier SM, Bui HH, Siddiqui J, Jing X, Cao X, Dhanasekaran SM, Feng FY, Chinnaiyan AM, Malik R. Identification and validation of PCAT14 as prognostic biomarker in prostate cancer. Neoplasia 2016;18:489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou J, Li M, Yu W, Li W, Wang J, Xiang X, Li G, Pan X, Lei D. AB209630, a long non-coding RNA decreased expression in hypopharyngeal squamous cell carcinoma, influences proliferation, invasion, metastasis, and survival. Oncotarget 2016;7:14628–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–310. [DOI] [PubMed] [Google Scholar]
- 18. Li W, Jia G, Qu Y, Du Q, Liu B, Liu B. Long Non-Coding RNA (LncRNA) HOXA11-AS promotes breast cancer invasion and metastasis by regulating epithelial-mesenchymal transition. Med Sci Monit. 2017;23:3393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen JH, Zhou LY, Xu S, Zheng YL, Wan YF, Hu CP. Overexpression of lncRNA HOXA11-AS promotes cell epithelial–mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int. 2017;17:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yao Y, Chen X, Lu S, Zhou C, Xu G, Yan Z, Yang J, Yu T, Chen W, Qian Y, Ding S, Tang J, Chen Y, Zhang Y. Circulating long noncoding RNAs as biomarkers for predicting head and neck squamous cell carcinoma. Cell Physiol Biochem. 2018;50:1429–40. [DOI] [PubMed] [Google Scholar]
- 21. Liu Z, Chen Z, Fan R, Jiang B, Chen X, Chen Q, Nie F, Lu K, Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer 2017;16:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H, Wang W, Li N, Li P, Liu M, Pan J, Wang D, Li J, Xiong Y, Xia L. LncRNA DGCR5 suppresses neuronal apoptosis to improve acute spinal cord injury through targeting PRDM5. Cell Cycle 2018;17:1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song W, Sun Y, Lin J, Bi X. Current research on head and neck cancer-associated long noncoding RNAs. Oncotarget 2018;9:1403–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu CZ, Jiang C, Wu Q, Liu L, Yan X, Shi R. A feed-forward regulatory loop between HuR and the long noncoding RNA HOTAIR promotes head and neck squamous cell carcinoma progression and metastasis. Cell Physiol Biochem. 2016;40:1039–51. [DOI] [PubMed] [Google Scholar]
- 26. Zhang TH, Liang LZ, Liu XL, Wu JN, Su K, Chen JY, Zheng QY, Huang HZ, Liao GQ. Long non-coding RNA MALAT1 interacts with miR-124 and modulates tongue cancer growth by targeting JAG1. Oncol Rep. 2017;37:2087–94. [DOI] [PubMed] [Google Scholar]
- 27. Fang Z, Zhang S, Wang Y, Shen S, Wang F, Hao Y, Li Y, Zhang B, Zhou Y, Yang H. Long non-coding RNA MALAT-1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins. BMC Cancer 2016;16:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, Wu Y, Mei M, Zhang L, Wang X. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial–mesenchymal transition in oral squamous cell carcinoma. Sci Rep. 2015;5:15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yim GW, Kim HJ, Kim LK, Kim SW, Kim S, Nam EJ, Kim YT. Long non-coding RNA HOXA11 antisense promotes cell proliferation and invasion and predicts patient prognosis in serous ovarian cancer. Cancer Res Treat. 2017;49:656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qu L, Jin M, Yang L, Sun C, Wang P, Li Y, Tian L, Liu M, Sun Y. Expression of long non-coding RNA HOXA11-AS is correlated with progression of laryngeal squamous cell carcinoma. Am J Transl Res. 2018;10:573–80. [PMC free article] [PubMed] [Google Scholar]
- 31. Zhan M, He K, Xiao J, Liu F, Wang H, Xia Z, Duan X, Huang R, Li Y, He X, Yin H, Xiang G, Lu L. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J CellMol Med. 2018;22(8):3758–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mu S, Ai L, Fan F, Sun C, Hu Y. Prognostic and clinicopathological significance of long noncoding RNA HOXA11-AS expression in human solid tumors: A meta-analysis. Cancer Cell Int. 2018;18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Su JC, Hu XF. Long noncoding RNA HOXA11AS promotes cell proliferation and metastasis in human breast cancer. Mol Med Rep. 2017;16:4887–94. [DOI] [PubMed] [Google Scholar]
- 34. Xue JY, Huang C, Wang W, Li HB, Sun M, Xi M. HOXA11-AS: A novel regulator in human cancer proliferation and metastasis. Onco Targets Ther. 2018;11:4387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu CW, Zhou DD, Xie T, Hao JL, Pant OP, Lu CB, Liu XF. HOXA11 antisense long noncoding RNA (HOXA11-AS): A promising lncRNA in human cancers. Cancer Med. 2018;7:3792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong W, Liao Y, Du J. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer 2013;49:604–15. [DOI] [PubMed] [Google Scholar]
- 37. Chen J, Wang BC, Tang JH. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106:260–6. [DOI] [PubMed] [Google Scholar]
- 38. Zhang CM, Zhao J, Deng HY. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci. 2013;20:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lao G, Liu P, Wu Q, Zhang W, Liu Y, Yang L, Ma C. Mir-155 promotes cervical cancer cell proliferation through suppression of its target gene LKB1. Tumour Biol. 2014;35:11933–8. [DOI] [PubMed] [Google Scholar]
- 40. Lv H, Guo J, Li S, Jiang D. miR-155 inhibitor reduces the proliferation and migration in osteosarcoma MG-63 cells. Exp Ther Med. 2014;8:1575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun J, Shi H, Lai N, Liao K, Zhang S, Lu X. Overexpression of microRNA-155 predicts poor prognosis in glioma patients. Med Oncol. 2014;31:911. [DOI] [PubMed] [Google Scholar]
- 42. Li T, Yang J, Lv X, Liu K, Gao C, Xing Y, Xi T. miR-155 regulates the proliferation and cell cycle of colorectal carcinoma cells by targeting E2F2. Biotechnol Lett. 2014;36:1743–52. [DOI] [PubMed] [Google Scholar]
- 43. Zhu X, Wang Y, Sun Y, Zheng J, Zhu D. MiR-155 up-regulation by LMP1 DNA contributes to increased nasopharyngeal carcinoma cell proliferation and migration. Eur Arch Otorhinolaryngol. 2014;271:1939–45. [DOI] [PubMed] [Google Scholar]
- 44. Shi LJ, Zhang CY, Zhou ZT, Ma JY, Liu Y, Bao ZX, Jiang WW. MicroRNA-155 in oral squamous cell carcinoma: Overexpression, localization, and prognostic potential. Head Neck 2015;37:970–6. [DOI] [PubMed] [Google Scholar]
- 45. Rather MI, Nagashri MN, Swamy SS, Gopinath KS, Kumar A. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: Implications for cancer therapeutics. J Biol Chem. 2013;288:608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lerner C, Wemmert S, Bochen F, Kulas P, Linxweiler M, Hasenfus A, Heinzelmann J, Leidinger P, Backes C, Meese E, Urbschat S, Schick B. Characterization of miR-146a and miR-155 in blood, tissue and cell lines of head and neck squamous cell carcinoma patients and their impact on cell proliferation and migration. J Cancer Res Clin Oncol. 2016;142:757–66. [DOI] [PubMed] [Google Scholar]
- 47. Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M, Carinci F. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. 2010;23:1229–34. [DOI] [PubMed] [Google Scholar]
- 48. Ni YH, Huang XF, Wang ZY, Han W, Deng RZ, Mou YB, Ding L, Hou YY, Hu QG. Upregulation of a potential prognostic biomarker, miR-155, enhances cell proliferation in patients with oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117:227–33. [DOI] [PubMed] [Google Scholar]